Fig. 4.
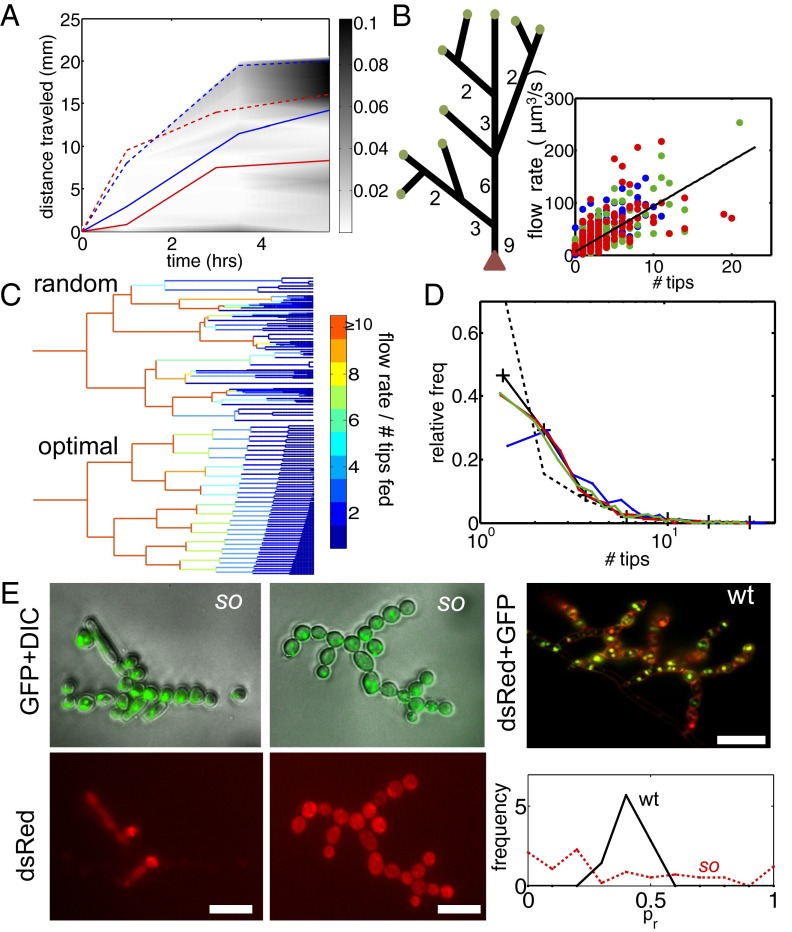
Mathematical models and the hyphal fusion mutant so reveal the separate contributions of hyphal branching and fusion to nuclear mixing. (A) pdf of distance traveled by nuclei entering a so colony. Mean (solid blue) and maximal (dashed blue) dispersal distances are similar to those of wild-type colonies (red curves, reproduced from Fig. 2B). (B) In so colonies, and <3 mm from the tips of a wild-type colony the network is tree-like, with a leading hypha (red arrowhead) feeding multiple tips (green circles). Hyphal flow rate is proportional to the number of tips fed so can be used to infer position in the branching hierarchy. (Inset) correlation of flow rate with number of tips fed in a real hyphal network. Blue, 3-cm colony; green, 4 cm; red, 5 cm 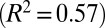 . (C) The probability
. (C) The probability 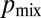 of sibling nuclei being sent to different tips was optimized by Monte Carlo simulations (SI Text). Optimal branching increases
of sibling nuclei being sent to different tips was optimized by Monte Carlo simulations (SI Text). Optimal branching increases 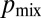 from 0.37 in a random branching network (Upper) to a value close to 0.46 (Lower). Branches are color coded by their flow rates. (D) For real colonies the distribution of branches at each stage of the hierarchy (blue, 3-cm mycelium; green, 4 cm; red, 5 cm) is close to optimal (solid black curve and crosses) rather than random branching (dashed black curve). (E) Despite having close to optimal branching, a so chimera becomes unmixed with growth. Conidial chains of a his-3::hH1-gfp; Pccg1-DsRed so + his-3::hH1-gfp; so heterokaryon tend to contain only hH1-GFP so nuclei (Left) or hH1-GFP DsRed so nuclei (Center); compare a heterokaryotic wild-type conidial chain in which hH1-DsRed and hH1-GFP nuclei are evenly mixed (Upper Right). (Scale bars, 20 μm.) Graph showing narrow spread of
from 0.37 in a random branching network (Upper) to a value close to 0.46 (Lower). Branches are color coded by their flow rates. (D) For real colonies the distribution of branches at each stage of the hierarchy (blue, 3-cm mycelium; green, 4 cm; red, 5 cm) is close to optimal (solid black curve and crosses) rather than random branching (dashed black curve). (E) Despite having close to optimal branching, a so chimera becomes unmixed with growth. Conidial chains of a his-3::hH1-gfp; Pccg1-DsRed so + his-3::hH1-gfp; so heterokaryon tend to contain only hH1-GFP so nuclei (Left) or hH1-GFP DsRed so nuclei (Center); compare a heterokaryotic wild-type conidial chain in which hH1-DsRed and hH1-GFP nuclei are evenly mixed (Upper Right). (Scale bars, 20 μm.) Graph showing narrow spread of 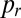 between wild-type conidial chains (black line) indicates more mixing of nucleotypes than in so (dashed red line).
between wild-type conidial chains (black line) indicates more mixing of nucleotypes than in so (dashed red line).