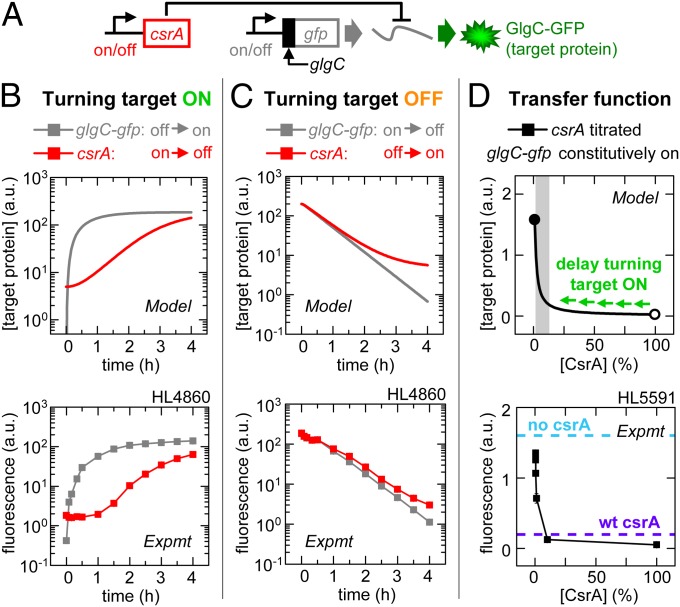
Fig. 2.
CsrA signaling: Stable signaling molecules can cause delays. Error bars are SEM of duplicate measurements. (A) Experimental schematic. (B) Target expression turned on directly by turning on glgC-gfp transcription (IPTG added) or indirectly by turning off csrA transcription (aTc removed). (C) Target expression turned off directly by turning off glgC-gfp transcription (IPTG removed) or indirectly by turning on csrA transcription (aTc added). (D) GlgC-GFP expression as a function of percentage maximum csrA transcription (calibrated using PLlacO-1:st7:gfp; Fig. S2Q). Target expression was also measured in strains without csrA (HL5594; cyan dashed line) or native (wt) csrA (HL5562 and HL5596; purple dashed line indicates both as the data overlay). The gray shading indicates the range over which the CsrA concentration has a significant effect on target expression (“regulatory range”). The open and closed circles are 100% and 0% of maximum [CsrA], respectively.