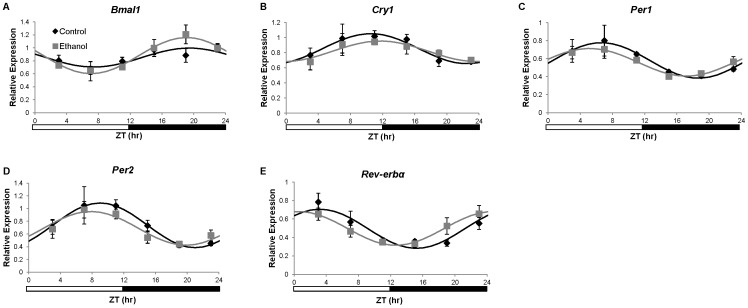
Figure 3. Expression of clock genes in the suprachiasmatic nucleus (SCN) of C57BL/6J mice.
Gene expression of a) Bmal1, b) Cry1, c) Per1, d) Per2, and e) Rev-erbα in SCN from mice fed control (♦) or ethanol-containing (grey square) liquid diets was determined using real-time quantitative PCR. Expression levels were normalized to the Gapdh housekeeping gene and are displayed as fold-change from a single control mouse. Cosinor analysis for curve fitting was performed using the nonlinear regression module in SPSS. Cosinor analysis results are included in Table 2. There were no statistically significant differences between the two feeding groups in mesor or amplitude in any genes examined in the SCN. Data are presented as mean ± SEM for n = 4 control and ethanol-fed mice at each time point.