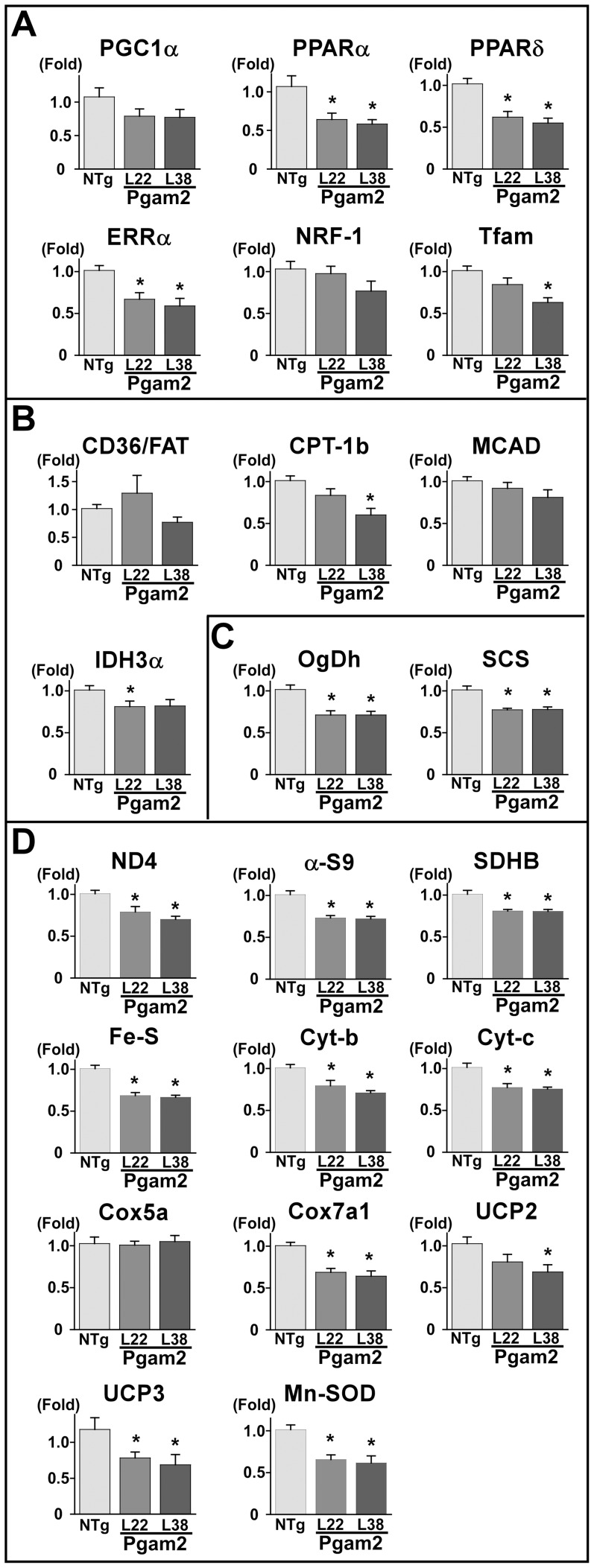
Figure 6. The expression of genes related to mitochondria was decreased in Pgam2 mice.
The expression of genes related to mitochondria was analyzed using quantitative real-time PCR. The genes presented here are involved in (A) transcriptional regulators, (B) fatty acid metabolism, (C) the TCA cycle, and (D) mitochondria. Peroxisome proliferator-activated receptor α (PPARα), peroxisome proliferator-activated receptor δ (PPARδ), estrogen related receptor α (ERRα), mitochondrial transcription factor A (Tfam), carnitine palmitoyltransferase 1b (CPT-1b), isocitrate dehydrogenase 3 α (IDH3α), oxoglutarate dehydrogenase (OgDh), succinyl-CoA synthetase α (SCS), mitochondrially encoded NADH dehydrogenase 4 (ND4), alpha-subcomplex 9 of complex I (α-s9), mitochondrial succinate dehydrogenase iron-sulfur subunit (SDHB), iron-sulfur protein (Fe-S), cytochrome b (Cyt-b), cytochrome c (Cyt-c), cytochrome c oxidase subunit VIIa (COX7a), uncoupling protein 2 (UCP2), uncoupling protein 3 (UCP3), and manganese superoxide dismutase (Mn-SOD) levels were decreased in Pgam2 mice. NRF-1: nuclear respiratory factor 1; CD36/FAT: CD36/fatty acid translocase; MCAD: medium-chain acyl coenzyme A dehydrogenase; Cox5a: mitochondrial cytochrome c oxidase subunit Va. The amount of target gene mRNA was normalized by 18S rRNA mRNA. Values are the mean ± SEM. Gene expression levels in Pgam2 mice were compared with those of NTg mice. *p<0.05 versus NTg mice (n = 12 for each group).