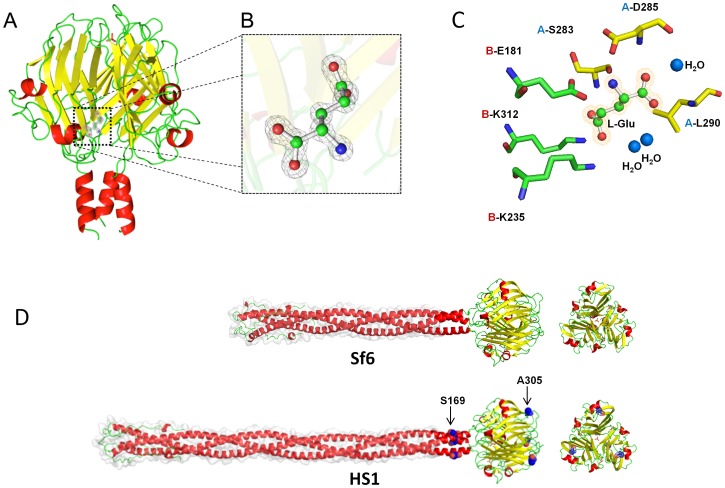
Figure 2. Atomic structure of the phage HS1 tail needle knob.
A. Ribbon diagram of bacteriophage HS1 tail needle knob determined crystallographically to 1.1 Å resolution; the N-termini are at the bottom of the diagram. Helices are shown in red, ß-sheets in yellow, and random coil in green; the bound L-glutamate is shown as sticks-and-balls and phosphate is shown as a small red sticks. B. Magnified view of L-glutamate trapped at the HS1 needle knob dimeric protomer:protomer interface. L-glutamate (in stick-and-balls) is overlaid to the final 2Fo-Fc electron density map (gray) contoured at 1.5σ above background. C. Side chains (sticks) from protomer A (yellow) and protomer B (green) that interact with L-glutamate (stick-and-balls). The indicated HS1 needle amino acids correspond to Sf6 needle amino acids as follows with Sf6 residue numbers in parentheses: Glu181(146), Lys235(200), Ser283(248), Asp285(250), Leu290(255) and Lys312(277). D. Structural models of full length Sf6 and HS1 tail needles. The two amino acid differences (from the Sf6 needle) that lie at positions in or near the knob domain, Ser169 and Ala305 of the HS1 tail needle, are shown as blue spheres. The models were obtained by using the Robetta full-chain protein structure prediction server [81]; the N-terminal parts of the needle protein shafts whose structures are modeled from the homologous P22 tail needle have a light gray surface contour behind. In all the panels, α-helices, β-strands and loops are colored in red, yellow and green, respectively.