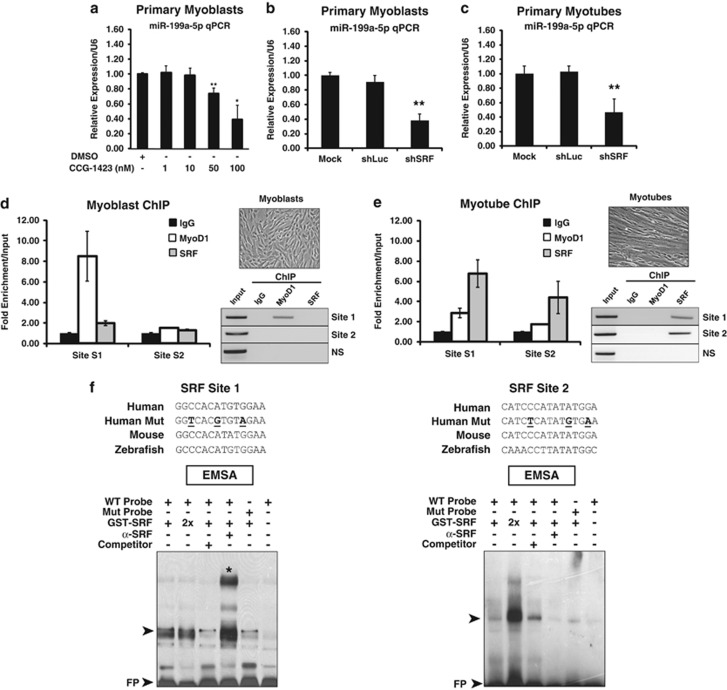
Figure 3.
SRF expression is essential for inducing miR-199a-5p expression in MB and MT. (a) Pharmacological inhibition of SRF blocks miR-199a-5p transcription levels. Real-time qPCR of miR-199a-5p levels of primary human MB treated with increasing levels of the SRF/Rho small-molecule inhibitor CCG-1423 as compared with a 10-nM DMSO control-treated cells. (b and c) SRF knockdown by shRNAi in MB (b) and MT (c) inhibits levels of miR-199a-5p transcript. A non-targeting (shLuciferase) construct is used as a negative control. *P-value<0.005; **P-value<0.05. (d and e) ChIP analysis of primary MB (d) and MT (e) reveals that SRF binds to the upstream (Sites 1 and 2) CArG boxes in differentiated human MT but not in MB (shown in upper panel). MyoD ChIP is used as a positive control in the myoblast samples. Real-time qPCR samples are normalized to input levels and showed as fold enrichment normalized to 10% input. A non-specific (ns) genomic sequence was also used as a negative control. ChIP PCR products obtained from MB and MT (both phase images shown) were resolved on agarose gels. All real-time qPCR of ChIP products were performed in triplicate. (f) EMSA of SRF protein binding CArG box Sites 1 and 2. Evolutionarily conserved putative CArG box sequence alignments are shown along with the three mutated nucleotides (bold, underlined). A stronger preference for Site 1 is observed. The top arrowhead denotes protein (SRF):DNA complex formation. A non-radiolabeled (competitor) was used to inhibit formation of the protein:DNA complex. The asterisk demarcates the supershift (SS) band. FP, free probe