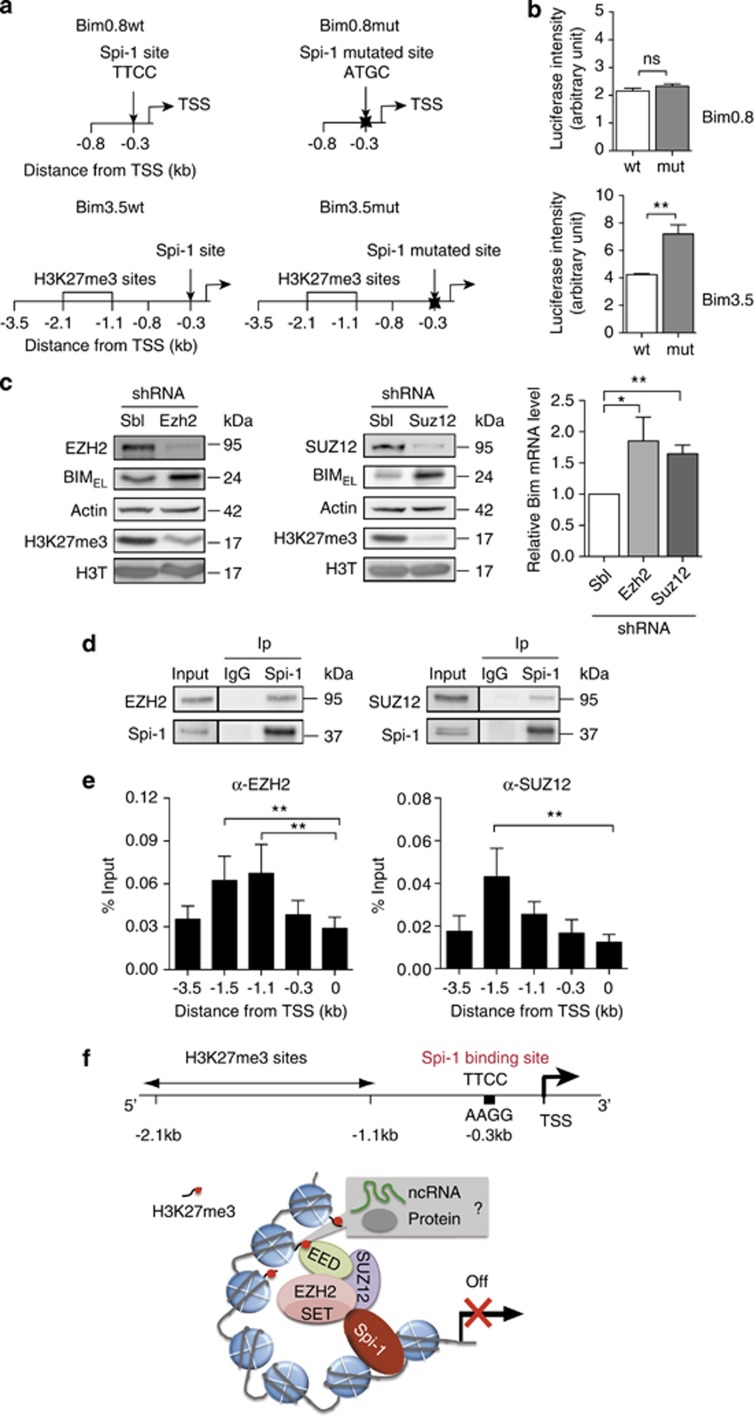
Figure 7.
Both Spi-1 binding to the Bim promoter and H3K27 trimethylation by PRC2 are required for the repression of Bim transcription. (a) Schematic representation of the Bim promoter–luciferase reporter constructs. Two fragments (0.8 and 3.5 kb) of the mouse Bim promoter were cloned in the pGL4 reporter vector to generate, respectively, the Bim0.8wt and Bim3.5wt vectors. The Spi-1 binding site (underlined) in the two vectors was then mutated (from 5′-AGTTCCGC-3′ to 5′-AGATGGGC-3′) to generate the Bim0.8mut and Bim3.5mut vectors. The TSS, the Spi-1 DNA binding site and the H3K27me3 sites are indicated. (b) shSpi-A2B cells were stably transfected with the different vectors (puromycin resistance) and luciferase assays were performed using exponentially growing cells 24 h after seeding. Data are reported as the relative firefly luciferase activity normalized to the number of cells and they represent the mean±S.D. of three experiments. Statistical differences were assessed by using the Student's t-test: **P<0.01. (c) shSpi-1-A2C cells were stably infected using lentiviral vectors encoding shRNAs against Ezh2 (shEZH2), Suz12 (shSUZ12) or control shSbl. The expression of EZH2, SUZ12, BIM, H3K27me3 and Actin was assessed by western blot (left) and the expression of Bim mRNA by real-time qPCR (right) in drug-selected cells. Bim mRNA level was normalized to Polr2α mRNA and to the level in shSbl. The bars represent the mean±S.E.M. of at least three independent experiments. Statistical analysis of the ΔCt values was carried out with the Student's t-test; **P<0.01 and *P<0.05. (d) Lysates from shSpi-1-A2B and shSpi-1-A2C cells were immunoprecipitated (Ip) with an anti-Spi-1 antibody or immunoglobulin G (IgG) as indicated. Immunoprecipitates were analysed by immunoblotting with anti-EZH2, anti-SUZ12 and anti-Spi-1 antibodies. (e) Chromatin isolated from shSpi-1-A2C or shSpi-1-A2B cells was IP using anti-EZH2 or anti-SUZ12 antibodies. The histogram bars represent the enrichment relative to the Input (%Input) determined by real-time qPCR using the indicated Bim promoter primers. The bars represent the mean±S.E.M. of three independent experiments. **P<0.01 by one-way analysis of variance (ANOVA) model followed by the post hoc test (pairwise t-test adjusted for multiple tests). (f) Working hypothesis for repression of Bim transcription by Spi-1. Spi-1 and PRC2 activities are necessary to favour trimethylation of H3K27 and to repress Bim transcription. The binding of Spi-1 and PRC2 to DNA occurs at distinct locations into the Bim promoter. These data indicate that Spi-1 promotes PRC2 activity without directly recruiting the complex to the site of its activity on the chromatin. Non-coding RNAs (ncRNAs) or other proteins may serve as intermediates in the PRC2 targeting at the methylation site and Spi-1 may act on these intermediates through stabilizing their interactions with PRC2