Abstract
Leaf spot and blight disease was observed on two-year-old seedlings of Dendropanax morbifera (Korean name: Hwangchil tree) during July of 2008 in Jindo Island, Korea. Symptoms included yellow-brown to dark brown irregularly enlarged spots frequently located along the veins of leaves. The lesions were often surrounded by chlorotic haloes. Severe leaf blight and subsequent defoliation occurred when conditions favored disease outbreak. The causal organism of the disease was identified as Alternaria panax based on morphological characteristics and sequence analysis of the internal transcribed spacer region of rDNA. A. panax isolates induced leaf spots and blight symptoms not only on D. morbifera but also on the other members of Araliaceae tested. This is the first report of foliar blight caused by A. panax on D. morbifera.
Keywords: Alternaria panax, Araliaceae, Dendropanax morbifera, Identification, Pathogenicity
Dendropanax morbifera Lev. is a subtropical, broadleaved evergreen tree belonging to the family Araliaceae. Lacquer derived from Dendropanax has a golden color and is an excellent varnish for use in woodworking and metalworking [1]. The lacquer is called Hwangchil in Korea, Koshiabura in Japan, and Jin-qi or Huang-qi in China [2]. Parts of D. morbifera can also be used in folk medicine for the treatment of migraine headaches and dysmenorrhea [3]. Further, D. morbifera is endemic to Korea but is naturally found only in the southern part [4, 5]. It has also been cultivated on Jindo Island, Suncheon, and Wando Island in Korea.
Severe leaf spots and blight disease of D. morbifera resulting in defoliation was observed on two-year-old seedlings in Jindo nurseries during July of 2008. A species of Alternaria was repeatedly isolated from diseased leaves of the plant and was suspected as the causal agent. However, the etiology of the disease has not been previously reported. The study reported here was initiated to determine the etiology of the disease.
Materials and Methods
Isolation of the fungus
Diseased leaves of Dendropanax morbifera were obtained from two-year-old seedlings in Jindo nurseries during July of 2008. Leaves with necrotic spots and blight symptoms were cut, placed in Petri dishes with moist filter paper, and incubated for 2~3 days at 25℃ to achieve sporulation of the causal organism. Alternaria isolates were obtained by single spore isolation from leaf spot symptoms and were deposited in the Culture Collection of Chungnam National University.
Morphological characteristics
Two isolates, CNU085017 and CNU085031, were used to determine morphology of the pathogen and were cultured on potato dextrose agar (PDA, Difco™, Detroit, MI, USA) for 3~5 days. Plugs (3 mm diameter) were taken from the edge of the colonies and transferred to PDA and V8 juice agar (V8A). After 7 days of incubation at 25℃, colony characteristics (color, growth rate, and pigment) were determined. Conidia produced on V8A (treated with 12/12 hr near-ultraviolet [NUV] light/darkness cycle) were mounted in lactophenol and measured using as light microscope (BX-50; Olympus, Tokyo, Japan) and Artray Artcam 300MI digital camera (Artray Inc., Tokyo, Japan). Identification of the fungus was conducted according to the description of Simmons [6, 7] and Yu [8].
DNA extraction, PCR amplification, and sequence analysis
The isolates were grown in potato dextrose broth for 5~7 days at 25℃. Mycelia were collected and freeze-dried for further extraction. Genomic DNA was extracted by the method described previously [9]. To identify the fungus, the internal transcribed spacer (ITS) region of rDNA was amplified using primers ITS5 (GGAAGTAAAAGTCGTAACAAGG) and ITS4 (TCCTCCGCTTATTGATATGC) [10]. PCR amplification of the ITS gene was performed in a 50-µL reaction mixture using a GeneAmp PCR System 2700 thermal cycler (Applied Biosystems, Foster City, CA, USA). The reaction was carried out with an initial denaturation for 3 min at 95℃, followed by 25 cycles of denaturation for 40 sec at 94℃, annealing for 1 min at 50℃, extension for 1 min at 72℃, and a final extension for 10 min at 72℃. PCR products were purified using a PCR Clean-up System purification kit (Promega, Madison, WI, USA) and sequenced using BigDye terminator cycle sequence kits (Applied Biosystems) with a ABI Prism 310 genetic analyzer (Applied Biosystems). The sequences of this fungus were compared with the ITS sequences of related Alternaria species available in the GenBank database by BLAST search. Sequences generated from the materials in this study and those retrieved from GenBank were initially aligned using the CLUSTAL X program [11], and the alignment was refined using PHYDIT program ver. 3.2 [12]. A neighborjoining tree was reconstructed with Kimura's 2-parameter distance model method [13] using PHYLIP 3.57c package [14]. Bootstrap analysis using 1,000 replications was performed to assess the relative stability of the branches.
Pathogenicity tests
Inoculation experiments for the two isolates of A. panax (CNU085017 and CNU085031) were carried out with detached leaves of D. morbifera and two species of Araliaceae (Aralia elata and Schefflera arboricola). Leaves of the non-host plant Glycine max were used as controls. Inocula for the experiments were grown in 9-cm-diameter Petri dishes containing V8A for 14 days at 25℃ under 12-hr photoperiods of NUV light. Sterile water (10 mL) was added to the plates, and the cultures were rubbed gently with a flamed glass rod. The resulting conidial suspensions were decanted into sterile test tubes and vigorously agitated with a vortex mixer for 30 sec before dilution and use. Young leaves were detached from each plant and surface-sterilized with 70% ethanol by gentle wiping. After the evaporation of ethanol, the leaves were inoculated at their center with 20 µL of conidial suspension (1 × 105 conidia/mL) and placed in plastic boxes with moistened paper towels. The boxes were sealed with parafilm and incubated at 22~25℃ for 7 days and then observed for symptom development.
Results and Discussion
Symptoms
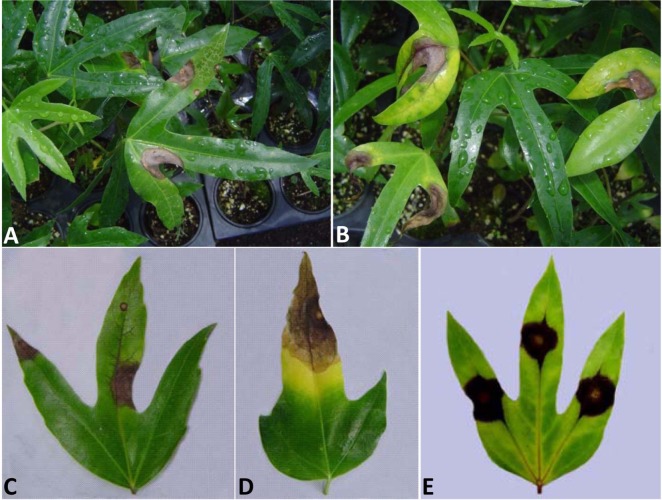
Initial symptoms on the leaves were observed as small, circular, and water-soaked spots. The spots were yellow-brown in the center with a broad dark brown border. The spots enlarged into lesions with irregular patterns frequently along the veins. A chlorotic halo often surrounded the lesions (Fig. 1A~1D). Severe leaf blight and subsequent defoliation occurred when the conditions favored disease outbreak.
Fig. 1.
Symptoms of foliar spots and blight on Dendropanax morbifera caused by Alternaria panax. A, B, Symptoms on naturally infected two-year-old seedlings; C, D, Lesions often advanced irregularly along with veins and were surrounded by a yellow halo; E, Symptoms on artificially induced leaves after 7 days of inoculation.
Morphological characteristics
Conidia of large-spored Alternaria sp. were abundantly produced on diseased leaves of D. morbifera incubated in moist chambers. Mostly large conidia with long beak were borne singly on the lesions, and others in chains of 2 to 3 formed in the moist chamber. Conidiophores were solitary or in clusters. The fungus can be isolated easily by selecting a single spore from the infected host material kept 2~3 days in a moist chamber.
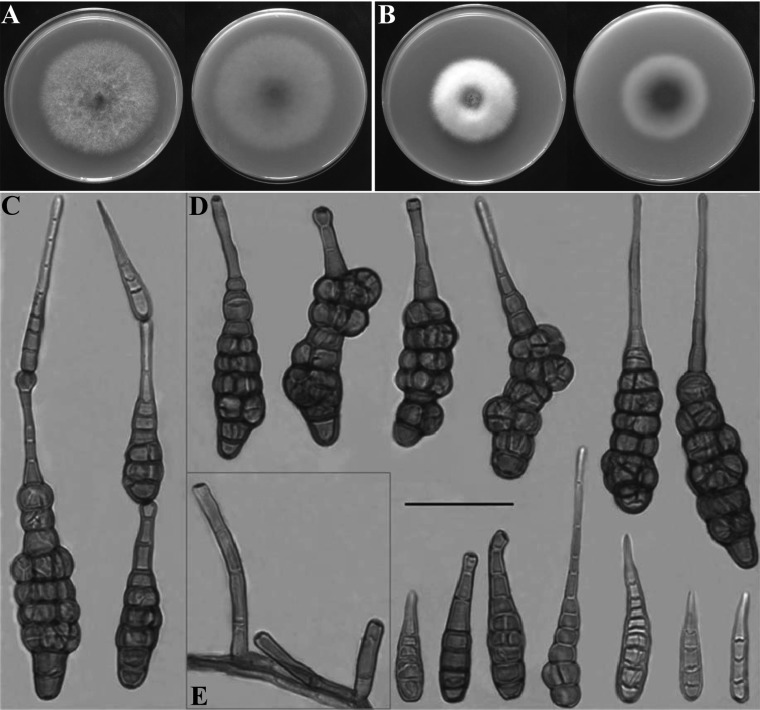
Colonies were velvety, pale gray, 60~65 mm in diameter on V8A (Fig. 2A) and were cottony, white, 45~50 mm in diameter on PDA after 7 days at 25℃. The fungus produced diffusible yellow pigment in the PDA substrate.
Fig. 2.
Morphology of Alternaria panax from Dendropanax morbifera. Colonies on V8 juice agar (V8A) (A) and potato dextrose agar (B) after 7 days of incubation at 25℃; Conidia (C, D) and conidiophores (E) of the fungus produced on V8A (scale bar = 50 µm).
Conidia sporulated abundantly on V8A under a 12/12 hr NUV light/darkness cycle (Fig. 2C and 2D). Conidiophores arose singly, terminally or laterally from hyphae, straight or curved, smooth-walled, septate, pale brown, usually with only one pigmented terminal conidiogenous site, slightly swollen at the apex, up to 150 µm long, 4~9 µm wide (Fig. 2E). Conidia solitary or 2~3 in small chains, straight or slightly curved, long elliptical or obclavate, goldenbrown to dark brown in color, smooth-walled when juvenile, becoming verruculose to verrucose, excessive cellular swelling causing the conidium to be distorted, with 5~11 transverse septa and 1 to several longitudinal or oblique septa, slightly or strongly constricted at the transverse septa, conidial body 70~120 × 15~30 (~45) µm. Some conidia had a beak (rostrate), some had a secondary conidiophore (pseudorostrate). Rostra almost cylindrical, rigid, simple, and unbranched, blunt-tipped, concolorous with conidium body or slightly lighter than the body, variable in length up to 83 µm, 3~7 µm wide. Pseudorostra cylindrical, simple, as short as 20 µm or as long as the spore body, 4~9 µm wide. These morphological characteristics agreed well with the Alternaria panax Whetzel as described by Simmons [6, 7] and Yu [8] (Table 1).
Table 1.
Comparison of morphological characteristics of Alternaria isolates from Dendropanax morbifera and previously described Alternaria panax
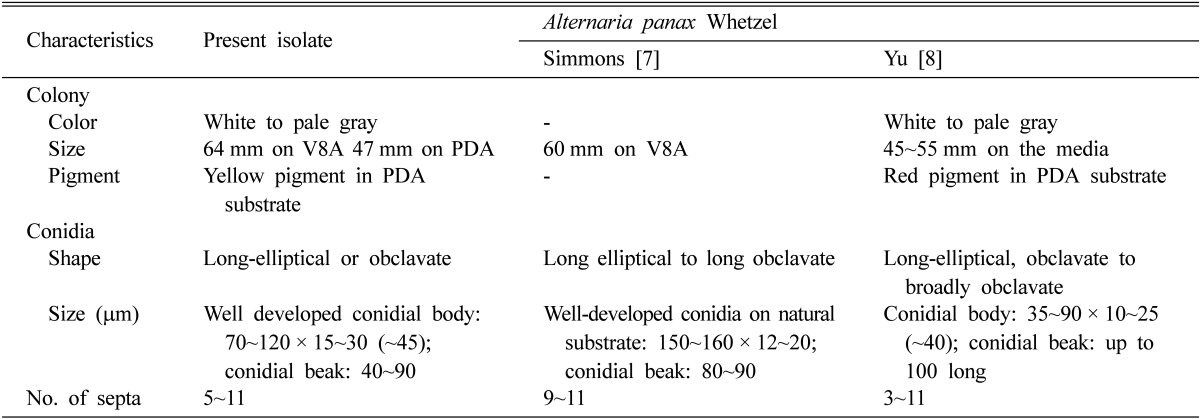
PDA, potato dextrose agar.
Phylogenetic analysis
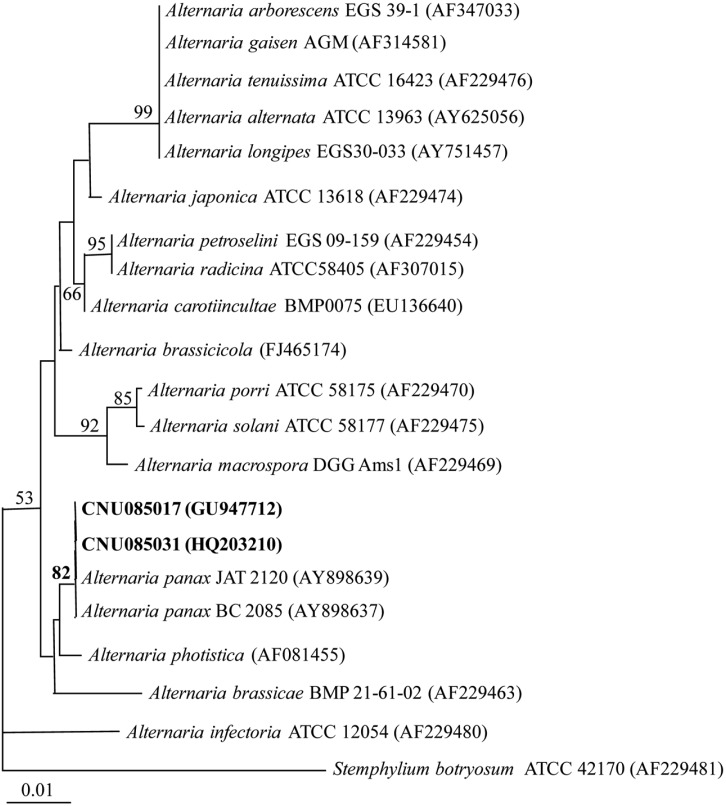
The ITS region sequences of CNU085017 and CNU085031 isolates of A. panax were deposited in GenBank (accession No. GU947712 and HQ203210). In a phylogenetic analysis using the neighbor-joining method, sequences of CNU085017 and CNU085031 isolates were found to be 100% identical to those of reference strains A. panax JAT 2120 and BC 2085 (GenBank accession No. AY898639 and AY898637), supported by a high bootstrap value (82%) (Fig. 3). They were also closely related to A. photistica (AF081455), as previously reported by Quayyum et al. [15]. The present phylogenetic analysis along with the morphological characteristics indicates that the causal fungus was A. panax.
Fig. 3.
Neighbor-joining tree based on sequence analysis of the internal transcribed spacer (ITS) region (ITS5/ITS4). Numbers close to the branch indicate bootstrap values obtained from 1,000 bootstrap replicates. Bar indicates the numbers of nucleotide substitutions per site. Accession numbers of GenBank are provided in parentheses.
Pathogenecity
The two isolates of the fungus were pathogenic to D. morbifera, Aralia elata, and Schefflera arboricola but not to G. max (Table 2). No significant differences were observed in pathogenicity between the two isolates, and the pathogen was frequently reisolated from the infected leaves. Symptoms appeared on D. morbifera leaves after 2~3 days of inoculation as small, dark, watersoaked spots. The spots enlarged into irregular lesions (Fig. 1E). These symptoms were identical to those observed on naturally infected plants.
Table 2.
Pathogenicity of Alternaria panax on detached leaves of three species of Araliaceae and Glycine max
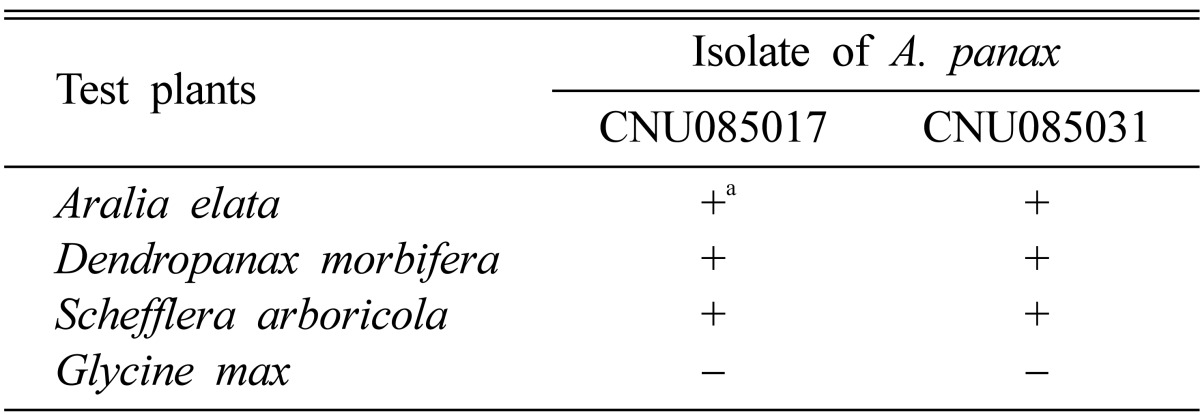
-, no symptoms developed; +, symptoms developed (lesion diameter > 15mm).
aLesion was observed at 7 days after inoculation.
To date, eleven hosts of A. panax have been reported [6-8, 16-18], and all are members of the Araliaceae family. These results provide the first documented evidence that D. morbifera is a new host of A. panax.
Acknowledgement
This work was supported by a grant from Regional Subgenebank Support Program of Rural Development Administration, Republic of Korea.
References
- 1.Kim SH, Jang YS, Han JG, Chung HG, Lee SW, Cho KJ. Genetic variation and population structure of Dendropanax morbifera Lev. (Araliaceae) in Korea. Silvae Genet. 2006;55:7–13. [Google Scholar]
- 2.Lee BH, Choi JH, Kim HJ. Curing and thermal behaviors of Korean Dendropanax Lacquer made by acetone and wine spirit extraction methods. Prog Org Coatings. 2005;52:241–245. [Google Scholar]
- 3.Bae KH. The medicinal plants of Korea. Seoul: Kyo-Hak Publishing Co Ltd.; 2000. [Google Scholar]
- 4.Han SH, Jung YH, Ko MH, Oh YS, Koh SC, Kim MH, Oh MY. Phytogenetic relationships of the Dendropanax morbifera and D. trifidus based on PCR-RAPD. Korean J Genet. 1998;20:173–181. [Google Scholar]
- 5.Kim SH. Ecology and superior tree selection of Dendropanax morbifera Lev [dissertation] Jinju: Gyeongsang National University; 1998. [Google Scholar]
- 6.Simmons EG. Alternaria: themes and variations. Mycotaxon. 1982;14:17–43. [Google Scholar]
- 7.Simmons EG. Alternaria: an identification manual. Utrecht: CBS Fungal Biodiversity Centre; 2007. p. 38. [Google Scholar]
- 8.Yu SH. Korean species of Alternaria and Stemphylium. Suwon: National Institute of Agricultural Science and Technology; 2001. pp. 100–106. [Google Scholar]
- 9.Park MS, Seo GS, Bae KS, Yu SH. Characterization of Trichoderma spp. associated with green mold of oyster mushroom by PCR-RFLP and sequence analysis of ITS regions of rDNA. Plant Pathol J. 2005;21:229–236. [Google Scholar]
- 10.White TJ, Bruns T, Lee S, Taylor JW. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. San Diego: Academic Press; 1990. pp. 315–322. [Google Scholar]
- 11.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chun J. Computer-assisted classification and identification of actinomycetes (Dissertation) Newcastle Upon Tyne (UK): University of Newcastle; 1995. [Google Scholar]
- 13.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 14.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 15.Quayyum HA, Dobinson KF, Traquair JA. Conidial morphology, virulence, molecular characterization, and host-parasite interactions of selected Alternaria panax isolates on American ginseng. Can J Bot. 2005;83:1133–1143. [Google Scholar]
- 16.Atilano RA. A foliar blight of Ming Aralia caused by Alternaria panax. Plant Dis. 1983;67:224–226. [Google Scholar]
- 17.Forsberg LI. Alternaria panax on umbrella tree. Australas Plant Pathol. 1983;12:7. [Google Scholar]
- 18.Garibaldi A, Gilardi G, Gullino ML. First report of Alternaria leaf blight of Aralia japonica caused by Alternaria panax in Europe. Plant Dis. 2004;88:82. doi: 10.1094/PDIS.2004.88.1.82B. [DOI] [PubMed] [Google Scholar]