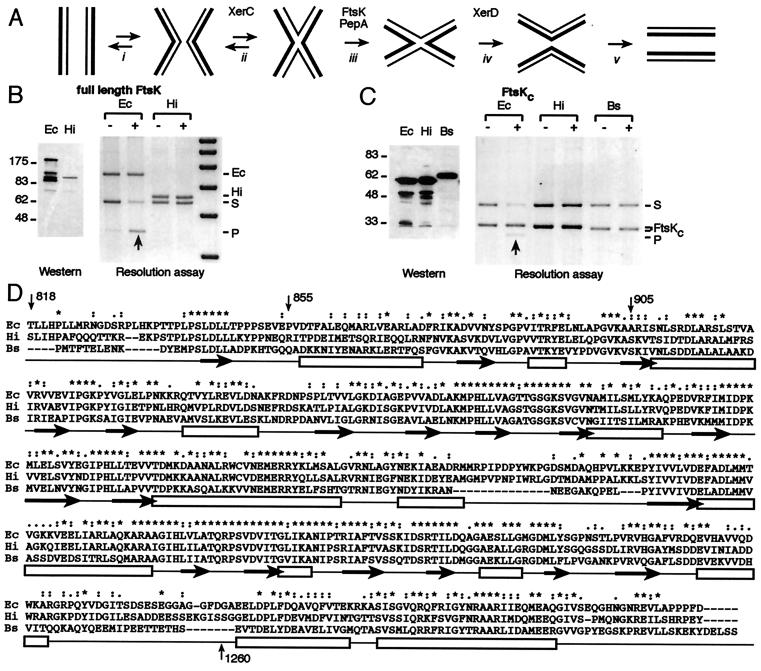
Figure 3.
(A) An outline of the Xer recombination reaction. XerCD bind cooperatively at dif, psi, or cer recombination sites, ensuring synapsis (with the help of accessory sequences and proteins in the case of psi/cer) (i). XerC initiates catalysis (ii) to form a HJ intermediate, which undergoes a conformational change (iii) to provide a substrate for catalysis by XerD, which can then complete the recombination reaction (iv). There is normally a barrier to this conformational change, and XerC frequently catalyzes the conversion of the HJ back to substrate (ii). In recombination at psi, the proteins PepA and ArgA-P facilitate the HJ conformational change, whereas in recombination at dif, FtsKc is thought to facilitate this change (34, 35). (B, C) Species specificity of FtsK action. FtsK cells (DS9041) were transformed with pBAD expression vectors (48) carrying full-length FtsK proteins (B) or the C-terminal domains (C) of different species. To assay for Xer recombination, they were transformed with a plasmid containing two dif sites and grown in conditions of repression (−; 0.2% glucose) or induction (+; 0.2% arabinose) of the expression vectors (34). Induction was checked by Western blot analysis by using an antibody directed against a FLAG epitope fused to the N termini of the constructs, after resolution of the protein extracts on a 6% (B) or an 8% (C) SDS/PAGE. (D) Alignment of the C-terminal domains of FtsK homologues. Identical residues are indicated by stars, conservative substitutions by dots. Open boxes underline regions predicted to adopt an α-helix conformation by predictprotein phd software v. 1.96, http://www.embl-heidelberg.de/predictprotein/predictprotein.html whereas black arrows underline those predicted to form β sheets. Ec: E. coli FtsK, Hi: H. influenzae FtsK and Bs: B. subtilis SpoIIIE.
cells (DS9041) were transformed with pBAD expression vectors (48) carrying full-length FtsK proteins (B) or the C-terminal domains (C) of different species. To assay for Xer recombination, they were transformed with a plasmid containing two dif sites and grown in conditions of repression (−; 0.2% glucose) or induction (+; 0.2% arabinose) of the expression vectors (34). Induction was checked by Western blot analysis by using an antibody directed against a FLAG epitope fused to the N termini of the constructs, after resolution of the protein extracts on a 6% (B) or an 8% (C) SDS/PAGE. (D) Alignment of the C-terminal domains of FtsK homologues. Identical residues are indicated by stars, conservative substitutions by dots. Open boxes underline regions predicted to adopt an α-helix conformation by predictprotein phd software v. 1.96, http://www.embl-heidelberg.de/predictprotein/predictprotein.html whereas black arrows underline those predicted to form β sheets. Ec: E. coli FtsK, Hi: H. influenzae FtsK and Bs: B. subtilis SpoIIIE.