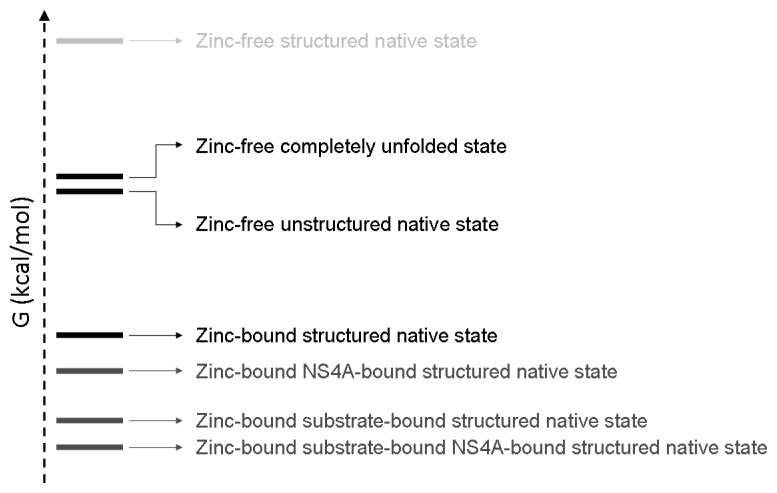
Figure 8.
Schematic depiction of the conformational landscape of NS3 protease at 20 °C considering its intrinsic structural stability and its interactions with its different ligands: zinc, NS4A and substrate. States are populated according to their Gibbs energy. Ligand (zinc, NS4A, substrate) binding modulates and shifts populations depending on the ligand binding affinity (and free ligand concentration, also). In the absence of zinc, the energetic gap between the fully unfolded state and the unstructured native state is very small (~0.4 kcal/mol), and the fully structured native state is hardly populated (high Gibbs energy). Binding of zinc, NS4A and substrate reduces the Gibbs energy of the protein. Because the binding affinity of the substrate is larger than that of NS4A ([25,62] and unpublished data), the binding of substrate stabilizes (lowers the Gibbs energy and increases the population) NS3 protease to a larger extent. States with very low population due to energetic penalty, such as the zinc-free NS4A-bound protease, are not shown.