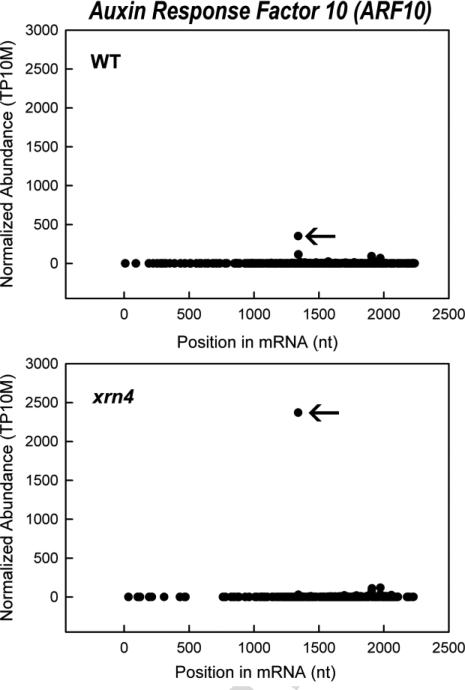
Fig 3. Example of decay plots (D-plots) showing accumulation of the downstream decay intermediate following miRNA-directed cleavage is elevated in an xrn4 mutant of Arabidopsis.
The abundance of PARE sequences are plotted versus their position within the ARF10 (AT2G28350.1) mRNA for the wild-type (WT) and loss-of-function mutant, xrn4. The arrows indicate the PARE sequence that corresponds to the position of miR160-directed cleavage. Accumulation of the downstream cleavage intermediate represented by this sequence is enhanced due to the absence of XRN4, which normally degrades this product. TP10M is transcripts per 10 million reads mapped.