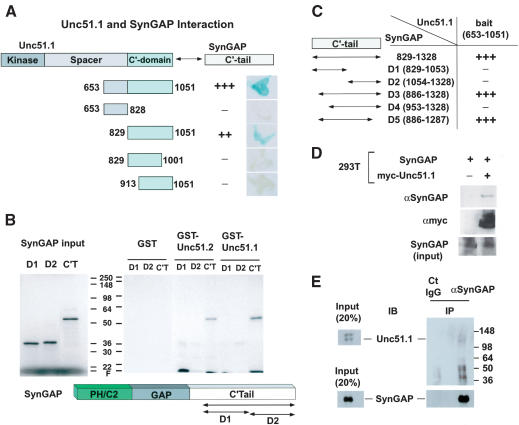
Figure 1.
Unc51.1 binds SynGAP. (A) Summary of the yeast two-hybrid assay. The C′-terminal half of Unc51.1 (amino acids 653–1051) and its deletion mutants were tested for their ability to bind SynGAP α2 C′-tail (amino acids 829–1328) in a β-galactosidase filter assay. Relative binding strength, as judged by darkness of the blue color of representative colonies, is indicated: (+++) very strong; (++) strong; (+) weak; (–) no binding detected. (B) GST pull-down assay. The C′-tail of SynGAP (amino acids 829–1328) and its deletion mutants D1 (amino acids 829–1053) and D2 (1054–1328) were in vitro translated, labeled with [35S]-methionine, and applied to the columns made with GST fusion proteins (GST alone, GST–Unc51.1 C′-tail [amino acids 653–1051], or GST–Unc51.2C′-tail [amino acids 531–1037]). Bound proteins were eluted from the columns and analyzed with SDS-PAGE, followed by autoradiography. (C) Determination of the minimum region of SynGAP that binds Unc51.1. Various deletion mutants (D1–D5) of the C′-tail of SynGAP were tested for their ability to bind Unc51.1 C′-terminal domain (amino acids 653–1051) in a β-galactosidase assay. (D) Coprecipitation assay. A full-length SynGAP expression construct was transfected into HEK293T cells with or without the myc-tagged full-length Unc51.1 construct. Cell extracts were immunoprecipitated with a myc antibody, and the immune complex was analyzed by immunoblot using an anti-SynGAP antibody. (E) Membrane-enriched fractions extracted from P6 cerebellum were immunoprecipitated with either the anti-SynGAP antibody or the normal rabbit IgG, and the resulting immune complexes were analyzed with SDS-PAGE followed by immunoblot with an anti-Unc51.1 antibody and an anti-SynGAP antibody.