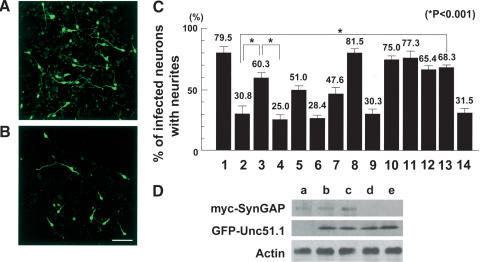
Figure 4.
SynGAP inhibition of cerebellar granule cell axon outgrowth and its rescue by Unc51.1. (A,B) Granule cells were purified from P6 cerebellum and infected with retroviruses expressing either GFP alone (A) or SynGAP + GFP (B), and axon outgrowth was visualized by a GFP antibody staining. Bar, 50 μm. (C) Quantitation of granule cells with neurites. Granule cells were infected with retroviruses expressing GFP alone (column 1), SynGAP + GFP (column 2), SynGAP + GFP–Unc51.1.wt (column 3), SynGAP + GFP–Unc51.1.KR (column 4), SynGAP + GFP–Unc51.1.wt/ΔC′-domain (column 5), SynGAP/ΔC′-tail + GFP (column 6), SynGAP/ΔC′-tail + GFP–Unc51.1.wt (column 7), GFP–Unc51.1.wt (column 8), GFP–Unc51.1.KR (column 9), SynGAP/Δ414-652+ GFP (column 10), GFP–H-Ras.wt (column 11), GFP–H-Ras17 (column 12), SynGAP + GFP–H-Ras.wt (column 13), or SynGAP + GFP–H-Ras17 (column 14), and the percentage of cells harboring neurites (>20 μm) were scored. A hundred cells per experiment were scored for the presence or absence of neurites and were counted from at least three individual experiments: 79.5% ± 6.2% (column 1), 30.8% ± 7.0% (column 2), 60.3% ± 4.5% (column 3), 25.0% ± 4.5% (column 4), 51.0% ± 2.7% (column 5), 28.4% ± 2.0% (column 6), 47.6% ± 6.1% (column 7), 81.5% ± 5.0% (column 8), 30.3% ± 5.4% (column 9), 75.0% ± 3.2% (column 10), 77.3% ± 5.5% (column 11), 65.4% ± 3.4% (column 12), 68.3% ± 1.8% (column 13) or 31.5% ± 4.8% (column 14) of cells expressing the indicated proteins extended neurites. Statistically significant differences (p < 0.001) were observed between columns 2 and 3, 3 and 4, and 2 and 13, as indicated by asterisks. (D) Expression levels of the proteins from the retroviruses used were evaluated. Equivalent titer (∼5 × 106 cfu) of each virus was used to infect NIH3T3 cells (∼1 × 106), and 48 h after the start of infection, cells were harvested and subjected for Western blotting by using myc and GFP antibodies. An actin antibody was used to ensure equal protein loading. Retroviruses used were myc-SynGAP + GFP (lane a), myc-SynGAP + GFP–Unc51.1.wt (lane b), myc-SynGAP + GFP–Unc51.1.KR (lane c), GFP–Unc51.1.wt (lane d), and GFP–Unc51.1.KR (lane e).