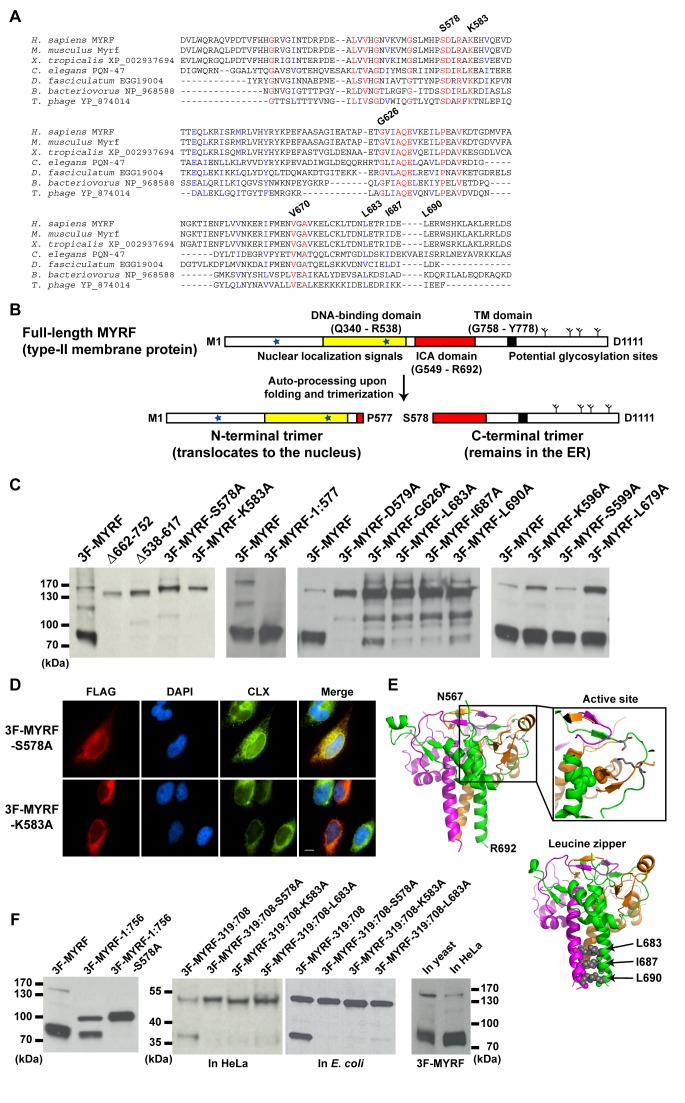
Figure 3. The ICA domain autonomously mediates the proteolytic processing of MYRF.
(A) Multiple sequence alignment of the ICA domains from eukaryotes, a bacterium, and a phage, generated with ClustalW [54]. Strictly conserved residues are shown in red. The numbering system is based on MYRF. (B) The auto-processing mechanism for MYRF postulated based on the ICA domain and its known properties. (C) Western blots of HeLa cells transfected with various MYRF constructs, showing the effects of mutations in the ICA domain on the proteolytic processing of MYRF. (D) IF image of 3F-MYRF-S578A and 3F-MYRF-K583A in HeLa cells. (E) The amino acid sequence of MYRF (residues N567-R692) was mapped onto the crystal structure of an ICA domain (PDB ID: 3GW6) using the alignment shown in Figure S3A. In the zoomed active site are shown two key catalytic residues (S578 and K583, both belonging to the same subunit) in stick model and two strictly conserved residues (V670 of one subunit and G626 of a different subunit) in space filling model. Shown below are L683, I687, and L690 that were predicted to form a leucine zipper. For visual clarity, clipped images were generated when deemed necessary. (F) (Left) Western blot showing that the proteolytic processing of MYRF is independent of its membrane insertion. MYRF-1:756 is a mutant truncated before the TM domain at L756. (Middle) Western blot showing the proteolytic processing of MYRF-319:708 in HeLa cells and E. coli. (Right) Western blot showing the normal processing of full-length MYRF in budding yeast. Scale bar, 10 µm.