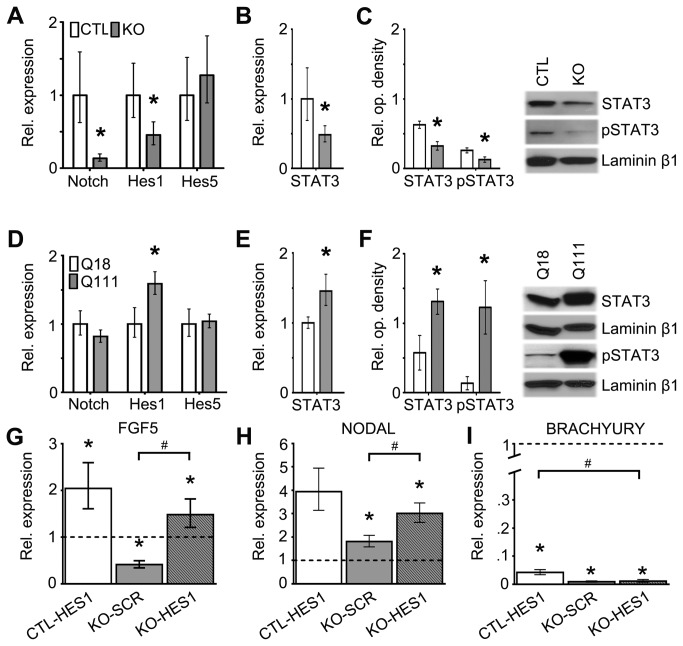
Figure 7. The regulation of Notch/Hes1/STAT3 signaling pathways requires Htt whereas mHtt differentially alters this process.
(A) QPCR expression analysis of Notch, Hes1 and Hes5 in CTL and KO EBs at 4DIV. (B) QPCR expression analysis of STAT3 in CTL and KO EBs at 4DIV. (C) Quantification of protein levels of non-phosphorylated and phosphorylated STAT3 measured by Western blot analysis in CTL and KO EBs at 4DIV. Error bars represent ± SEM; *p<0.05. (D) QPCR expression analysis of Notch, Hes1 and Hes5 in Q18 and Q111 EBs at 4DIV. (E) QPCR expression analysis of STAT3 in Q18 and Q111 EBs at 4DIV. (F) Quantification of protein levels of non-phosphorylated and phosphorylated STAT3 measured by Western blot analysis in Q18 and Q111 EBs. Error bars represent ± SEM; *p<0.05. (G–I) QPCR expression analysis of the germ layer markers, FGF5, NODAL and BRACHYURY in CTL and KO Hes1-overexpression EBs. Dotted lines refer to expression levels of control ESCs expressing the scrambled construct, CTL-SCR. Error bars represent ±95% CI. *p<0.0001 unless otherwise noted, as compared to CTL-SCR; # p<0.0001 unless otherwise noted, as compared to KO–SCR (G, H) and CTL-HES1 (I).