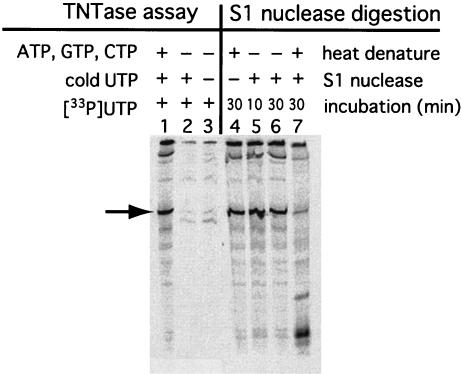
FIG. 4.
TNTase assay and S1 nuclease digestion. An in vitro-transcribed RNA, 606-polyA, corresponding to the 3′-terminal 232 nucleotides of the NV genome, was used as a template for the RdRp reaction. For TNTase assays, reactions were performed without ATP, GTP, and CTP (lane 2) or without ATP, GTP, CTP, and cold UTP (lane 3). An RNA product from a standard reaction mixture is shown (lane 1) as a control. For the S1 nuclease digestion analyses, RdRp products were heat denatured at 95°C for 2 min (lanes 4 and 7) before treatment without (lane 4) or with (lanes 5, 6, and 7) S1 nuclease for the indicated times. The 232-nucleotide band, identified by an arrow, is missing from lanes 2, 3, and 7. RNA products were analyzed by methods similar to those described for Fig. 2.