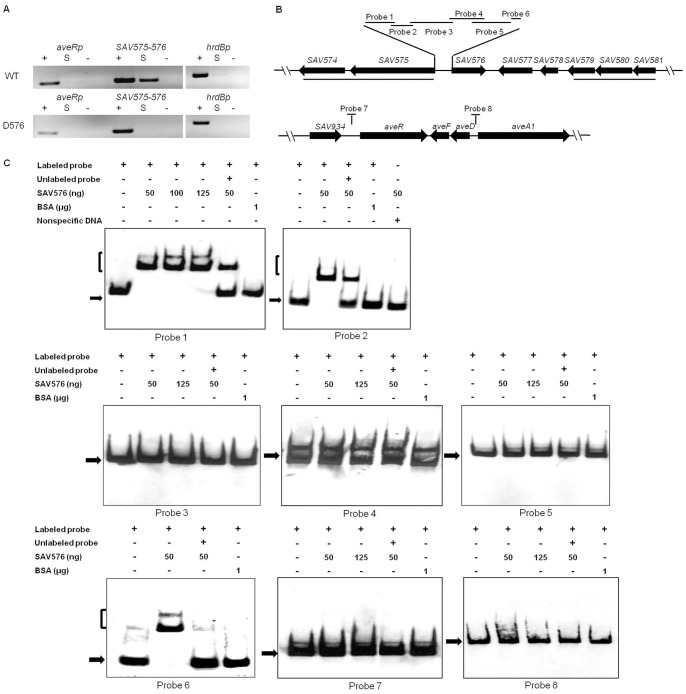
Figure 4. Analysis of SAV576 protein binding to target promoter regions.
(A) ChIP assays in vivo. Anti-SAV576 antibodies were used to immunoprecipitate SAV576-DNA complexes from ATCC31267 and D576 cells treated with formaldehyde. The DNAs used for PCR were total DNA prior to immunoprecipitation (positive control: lanes “+”), immunoprecipitated DNA (experimental sample: lanes “S”), and negative control DNA without antibody (lanes “−”). The hrdB promoter region was used as a control. (B) Schematic representation of the relative positions of probes used for EMSAs in vitro. Probe 1, 98-bp DNA fragment from +2 to −96 relative to the translational start codon of SAV575; probe 2, 43-bp fragment from −98 to −140; probe 3, 276-bp fragment from −118 to −393; probe 4, 257-bp fragment from −306 to −562; probe 5, 333-bp DNA fragment from −533 to −865; probe 6, 43-bp DNA fragment from −866 to −908. Probe 7, 200-bp DNA fragment from −116 to −315 relative to the start codon of aveR. Probe 8, 328-bp DNA fragment from −14 to −341 relative to the start codon of aveA1. Probes 7 and 8 cover the putative transcriptional start points of aveR and aveA1, respectively. (C) EMSAs of the interaction of the probes with purified His6-SAV576 protein. Each lane contained 0.3 nM labeled probe. The labeled probe and an approximately 100-fold excess of the unlabeled probe were used in competitive assays. BSA was used as a negative control for SAV576 protein. Labeled non-specific DNA was used to eliminate non-specific binding of SAV576 protein. The free probes are indicated by solid arrows, and the retarded DNA fragments are indicated by parentheses.