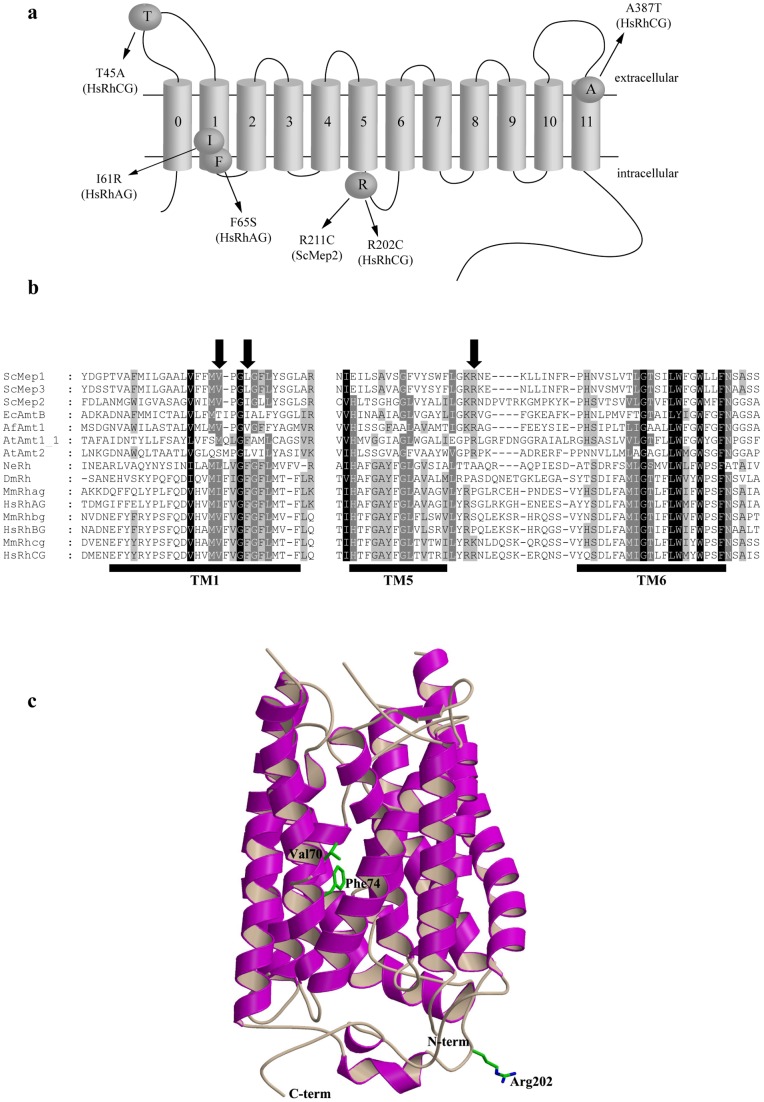
Figure 1. Location of the considered amino acid substitutions in HsRhAG, HsRhCG, and ScMep2.
a Schematic representation of the predicted topology of Mep-Amt-Rh proteins. The first transmembrane segment (0) is absent in ScMep2 protein. b Partial primary sequence alignment of Mep-Amt-Rh proteins from different organisms. SWISS PROT or GeneBank accession numbers are referred hereafter. ScMep1, 2, 3: S. cerevisiae Mep1 (P40260), Mep2 (P41948), and Mep3 (P53390); EcAmtB: E. coli AmtB (P69681); AfAmt1: A. fulgidus Amt1 (O29285); AtAmt1_1, 2: A. thaliana Amt1;1 (P54144) and Amt2 (Q9M6N7); NeRh: N. europaea Rh50 (Q82X47); DmRh: D. melanogaster Rh (Q9V3T3); MmRhag, bg, cg: M. musculus Rhag (Q9QUT0), Rhbg (Q8BUX5) and Rhcg (Q9QXP0); HsRhAG, BG, CG: H. sapiens RhAG (Q02094), RhBG (AAL05978) and RhCG (Q9UBD6). The limits of transmembrane helices (TM) 1, 5 and 6 of human HsRhCG are deduced from the crystal structure of this protein (pdb code 3HD6) and are underlined. The location of Ile61 and Phe65 of HsRhAG and Arg202 of HsRhCG is indicated by an arrow. c Ribbon representation of the HsRhCG X-ray structure with labelled and depicted Val70, Phe74 and Arg202 residues.