Figure 1. Flowchart for constructing human brain anatomical networks using DTI datasets and tractography.
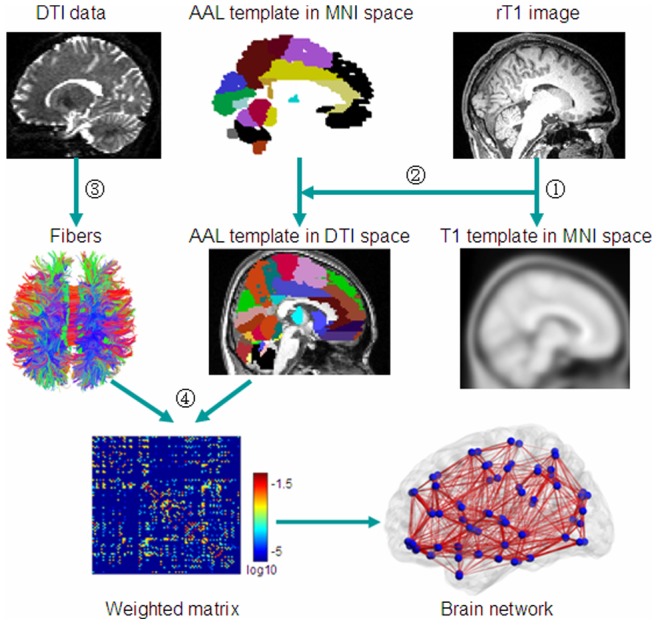
(1) Individual anatomical images were first coregistered into b0 images to obtain rT1 images in diffusion space. The rT1 images were then mapped to a T1-weighted template of ICBM152 in MNI space. (2) The obtained inverse matrix was used to transform the AAL template from the MNI space into individual diffusion space. (3) Fibers in the whole brain were reconstructed using the deterministic tractographic method (DtiStudio software). For display purposes only, fibers shown here were calculated using TrackVis software. (4) Construction of the weighted connectivity matrices and the human brain anatomical networks.
