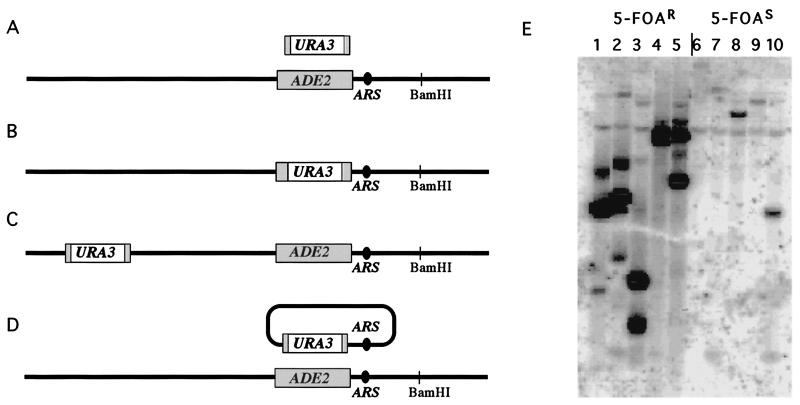
Figure 3.
(A) Gene targeting at the ADE2 locus using an URA3 gene carrying only 50 bp of ADE2 sequences on either end. (B) An ARS sequence lies just upstream of the ADE2 gene, which is transcribed right to left. Three types of transformants were recovered. About half had replaced ADE2 with URA3 sequences. (C) Another group had inserted the URA3 nonhomologously at different sites in the genome. (D) A third group had formed unstable, apparently circular autonomously replicating sequences in which the ADE2-adjacent ARS had been copied onto the DNA of the transforming fragment, presumably by a BIR-like event. (E) Examples of BamHI-digested DNA from 5-FOA-resistant (5-FOAR) colonies harboring an unstable replicating URA3 gene (lanes 1 to 5), which is present in high copy number when probed with URA3 sequences. There is no BamHI site within 5000 bp upstream and 2200 bp downstream of the ADE2 gene. Supercoiled, nicked circular and linearized forms are evident. In lanes 6–10 are 5-FOA-sensitive colonies where the URA3 gene had integrated at unknown locations, present at single copy.