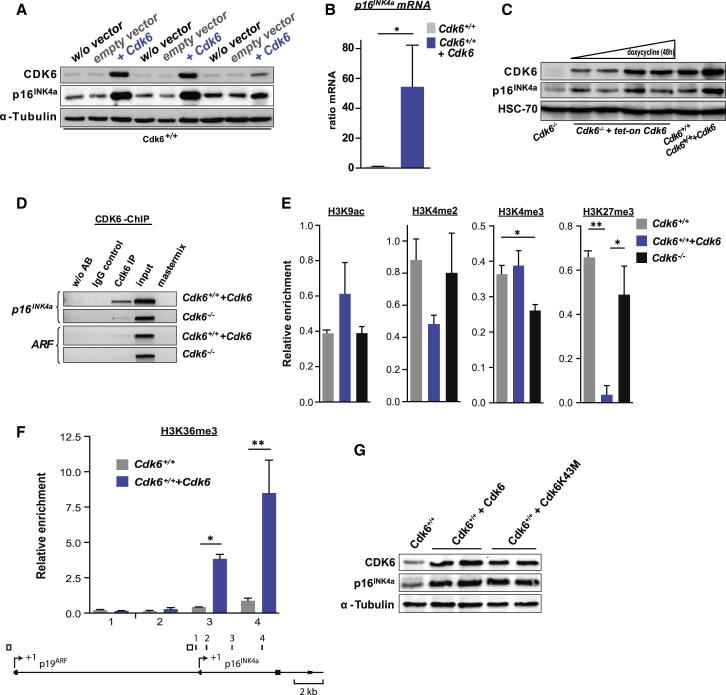
Figure 2.
CDK6 Acts as a Transcriptional Regulator of p16INK4a
(A) Immunoblot for CDK6 and p16INK4a of three individually derived Cdk6+/+ cell lines with (+Cdk6) or without (w/o vector, empty vector) enforced CDK6 expression.
(B) p16INK4a mRNA levels of Cdk6+/+ and Cdk6+/++Cdk6 cells were analyzed with qPCR (n = 6; ∗p = 0.03).
(C) Immunoblot for CDK6 and p16INK4a of Cdk6−/− cells expressing a doxycycline-inducible tet-on Cdk6 vector (Cdk6−/−+tet-on Cdk6). Lane 1: Cdk6−/− cells. Lanes 2–5: Cdk6−/−+tet-on Cdk6 cells 48 hr after treatment with doxycycline (0, 0.3, 1, 3 μM). Lane 6: Cdk6+/+ cells. Lane 7: Cdk6+/++Cdk6 cells.
(D) ChIP assays were performed using Cdk6+/++Cdk6 and Cdk6−/− cells. Protein-DNA complexes were immunoprecipitated using an anti-CDK6-antibody and analyzed by PCR for the presence of p16INK4a and as specificity control of ARF promoter sequence (region depicted in Figure 2F as open rectangles).
(E) Promoter ChIP assays were performed using Cdk6+/+, Cdk6+/++Cdk6, and Cdk6−/− cells. Protein-DNA complexes were immunoprecipitated using antibodies specific for the indicated histone modification. ChIP DNA was analyzed with qPCR for the presence of a p16INK4a promoter sequence (region 1 in Figure 2F). The relative enrichment to a control region is shown (Tbp promoter for H3K9ac, H3K4me2, and H3K4me3; Neurog1 promoter for H3K27me3). The mean and SEM of two independent experiments is shown (H3K4me3: Cdk6+/+ versus Cdk6−/−, ∗p < 0.05; H3K27me3: Cdk6+/+ versus Cdk6+/++Cdk6,∗∗p < 0.01; Cdk6+/++Cdk6 versus Cdk6−/−, ∗p < 0.05).
(F) H3K36me3 ChIP assays were performed using Cdk6+/+ and Cdk6+/++Cdk6 cells. Protein-DNA complexes were immunoprecipitated using an antibody specific for H3K36me3. ChIP DNA was analyzed with qPCR for the presence of p16INK4a sequences 1–4, which are depicted as filled black rectangles in the lower panel (middle of the amplicon relative to the TSS [arrow symbol marked +1] of p16INK4a: 1, −169 base pairs; 2, +538 base pairs; 3, +2,278 base pairs; 4, +4,312 base pairs). The relative enrichment to a Gapdh gene body region is shown. The mean and SEM of two independent experiments is shown (sequence 3: Cdk6+/+ versus Cdk6+/++Cdk6,∗p < 0.05; sequence 4: Cdk6+/+ versus Cdk6+/++Cdk6,∗∗p < 0.01).
(G) Immunoblot for CDK6 and p16INK4a of Cdk6+/+, Cdk6+/++Cdk6, and Cdk6+/++Cdk6K43M cells.
See also Figure S2.