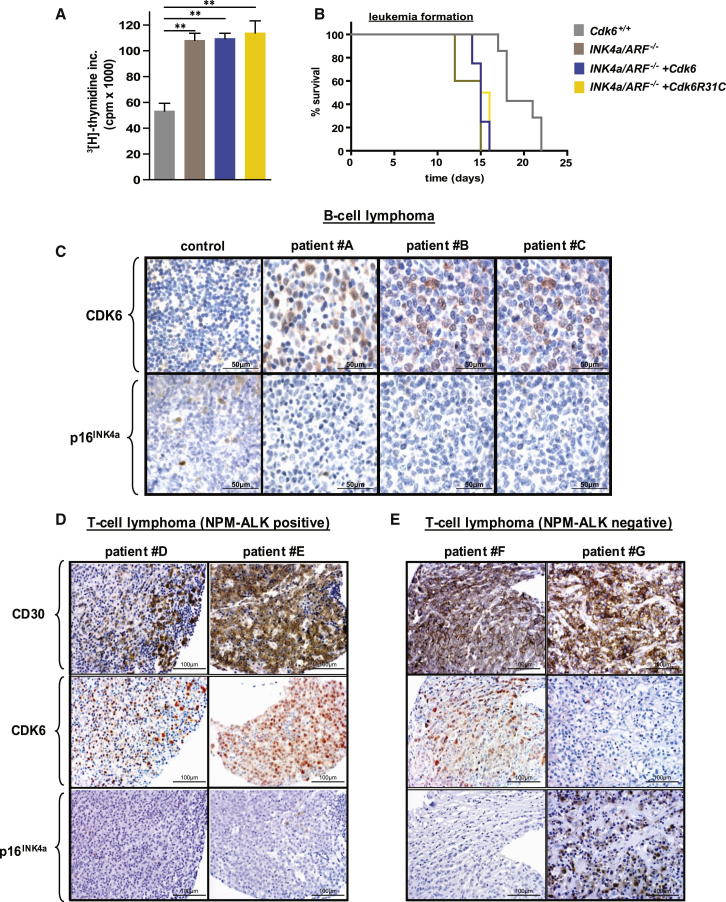
Figure 3.
Inverse Relation between CDK6 and p16INK4a Expression in Human Lymphomas
(A) [3H]thymidine incorporation of Cdk6+/+, INK4a/ARF−/−, INK4a/ARF−/−+Cdk6, and INK4a/ARF−/−+Cdk6R31C p185BCR-ABL-transformed cells (n = 3; Cdk6+/+ versus INK4a/ARF−/−, ∗∗p = 0.002; INK4a/ARF−/−+Cdk6, ∗∗p = 0.001; INK4a/ARF−/−+Cdk6R31C,∗∗p = 0.005).
(B) Kaplan-Meier plot of Rag2−/− mice intravenously transplanted with Cdk6+/+, INK4a/ARF−/−, INK4a/ARF−/−+Cdk6, and INK4a/ARF−/−+Cdk6R31C cells (n = 3 cell lines/genotype; mean survival: 18 [Cdk6+/+], 15 [INK4a/ARF−/−], 15 [INK4a/ARF−/−+Cdk6], and 15.5 [INK4a/ARF−/−+Cdk6R31C] days; ∗p = 0.02).
(C) Immunohistochemical stainings of a B cell lymphoma tissue array including 16 lymphoma samples and two control lymph nodes for CDK6 and p16INK4a. Representative examples including different types of B cell lymphoma, a diffuse large B cell lymphoma (patient A), two follicular lymphomas (patients B and C), and one control lymph node (left) are depicted. Original magnification 20×.
(D and E) The expression of CD30, CDK6, and p16INK4a was analyzed for 17 NPM-ALK-positive (E) and 11 NPM-ALK-negative (D) lymphoma cases with immunohistochemistry. Representative cases for NPM-ALK-positive (patients D and E) and NPM-ALK-negative (patients F and G) cases are depicted. Original magnification 20×.
Error bars indicate the mean ± SEM. See also Figure S3.