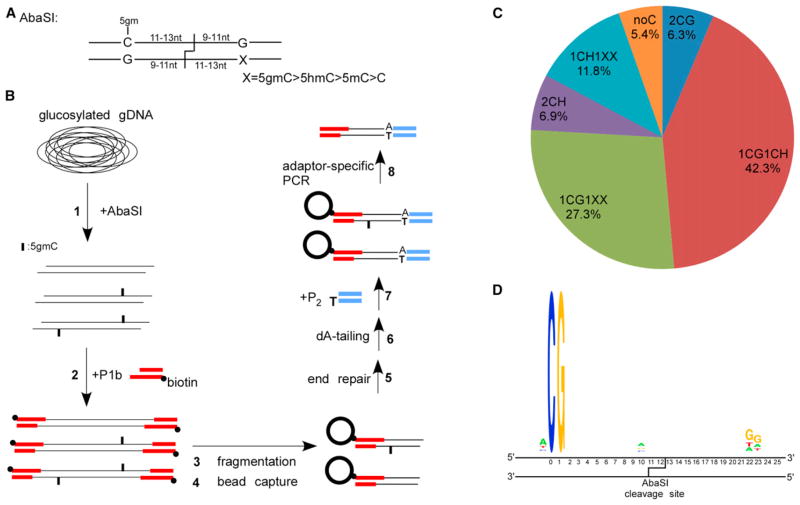
Figure 1. Overview of Aba-seq and Validation of the Identified 5hmC Sites.
(A) Recognition and cleavage sites of AbaSI.
(B) Schematic overview of the Aba-seq method. 1: glucosylated genomic DNA was digested with AbaSI. 2: digested DNA was ligated to a biotinylated adaptor P1b with 2-base 3′-overhang. 3: DNA fragmentation by sonication. 4: capture of the biotinylated DNA onto avidin-coated beads. 5 and 6: on-bead DNA end-repair and dA-tailing. 7: on-bead ligation with adaptor P2 containing one blunt end and one unligatable end. 8: PCR using primers specific to the P1b and P2.
(C) Distribution of different categories of identified AbaSI cleavage sites.
(D) Representative sequence logos of the largest categories 1CG1CH.