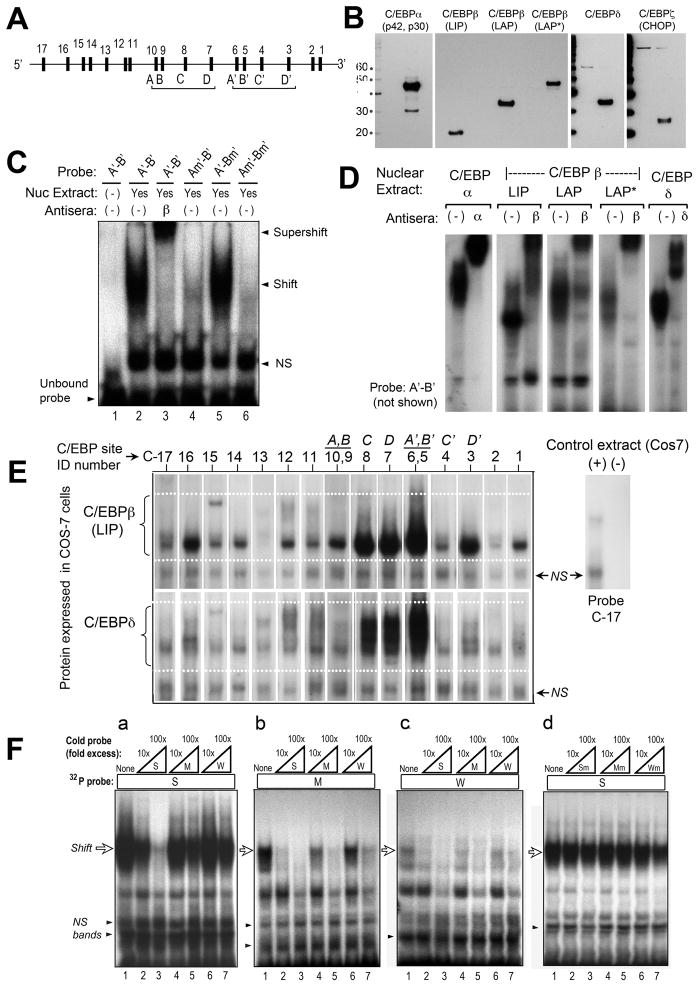
Figure 3. Characterization of putative C/EBP binding sites in the murine CPO promoter, by electrophoretic mobility shift analysis (EMSA).
(A) Schematic locations of the 17 candidate sites. (See Fig. 5a for exact locations.) (B) Western blots to verify proper expression of C/EBP isoforms. COS-7 cells were transfected with expression vectors encoding each of the C/EBP isoforms indicated, then analyzed on western blots using specific antisera. A 10 kDa molecular weight ladder was run in parallel, to establish the relative sizes of the visualized bands (kDa, on left). For the three C/EBPβ isoforms (LIP, LAP, and LAP*) which are translated from alternative translation start sites, a single antibody to the C-terminal end of C/EBPβ was used. (C) Demonstration of specific binding at site A’. Nuclear extracts containing C/EBPβ were incubated with the 32P-labeled double-stranded DNA probe indicated above each lane. Each probe contained two candidate sites (A’ and B’), located 16 bp apart. For probe Am’-B’ or A’-Bm’, the sites A’ or B’ (respectively) were disrupted by mutation. NS, nonspecific bands. (D) Binding of different C/EBP isoforms to site A’. The test probe, A’-B’, was incubated with nuclear extracts overexpressing different C/EBP isoforms. Antibodies to C/EBPα, C/EBPβ, or C/EBPδ were added in some of the binding reactions to demonstrate specificity of the DNA-protein complexes (supershifts). (E) EMSA to test the relative binding of C/EBP isoforms to candidate sites in the mCPO gene. EMSA was performed with double-stranded DNA probes representing each of the 17 candidate sites. In the figure, specific DNA-protein complexes formed with C/EBPβ-LIP (upper panel), or with C/EBPδ (lower panel), are illustrated. The results of experiments with all of the C/EBP isoforms are summarized in Table 1. (F) Cross-competition analysis to test a predicted hierarchy of binding affinities. Three binding probes with relative affinities of 40% (site C6), 6% (site C3), and 3% (site C10) were chosen and for simplicity renamed strong (S), medium (M), and weak (W), respectively. Binding of C/EBPβ-LAP to these 32P-labeled probes was tested by EMSA in the absence or presence of excess amounts (10-fold or 100-fold) of unlabeled probe, in order to assess the ability of unlabeled competitors to diminish 32P-probe binding. Subpanels a, b, and c: Unlabeled competitors were able to displace 32P-probe binding as predicted by the relative order of binding affinities predicted from Table 1. Subpanel d: mutated sites were unable to compete (negative controls).