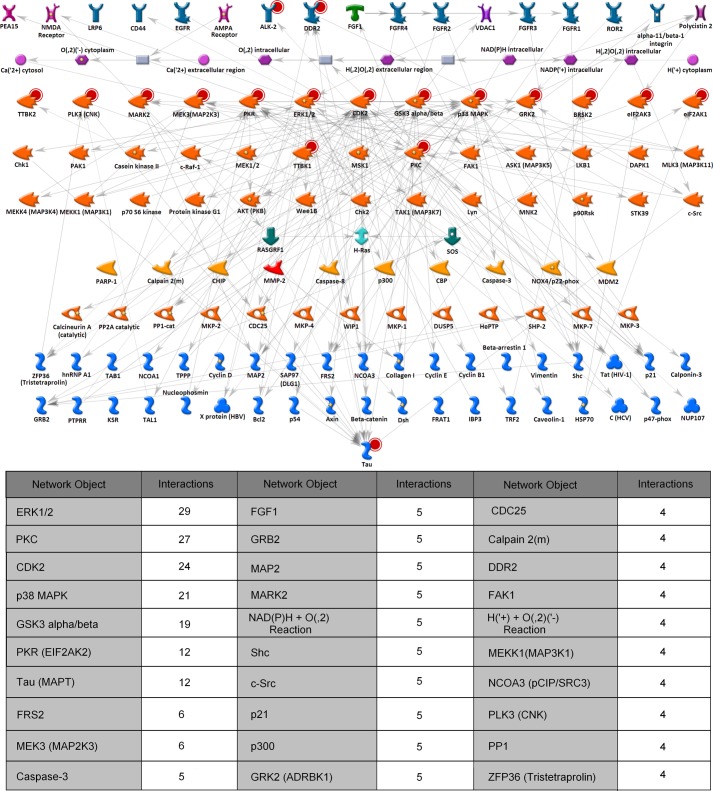
FIGURE 10.
Network fishing for molecules involved in tau phosphorylation pathways. The network shown was generated using the GeneGo MetaCore pathway analysis software to trace the putative pathways from and through the kinases identified in the kinase screen and to the tau protein. Red circles behind the icons show nodes used as input to the network building algorithm. For ease of representation, transcription factors were removed from the network, the nodes were organized by protein families, and the edges were grayed out. The table below the network shows some of the hubs in the network and the number of interactions made by each molecule. EGFR, EGF receptor; cat, catalytic; HBV, hepatitis B virus; HCV, hepatitis C virus; CBP, cAMP-response element-binding protein (CREB)-binding protein. CNK, connector enhancer of KSR; KSR, kinase suppressor of RAS; SOS, son of sevenless guanine nucleotide exchange factor; PTPRR, protein-tyrosine phosphatase receptor type R; HePTP, hematopoietic protein-tyrosine phosphatase.