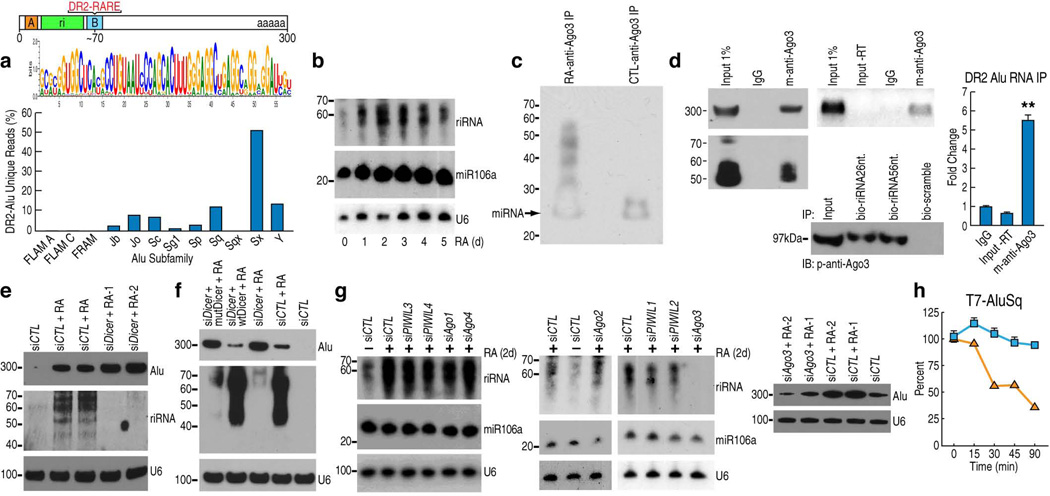
Figure 2.
DR2 Alu transcripts associate with AGO3 and produce riRNAs in a DICER-dependent manner. (a) Bioinformatics analysis of small RNA-seq datasets identified a consensus sequence of riRNAs derived from the 5’ end of DR2 Alu (the green box). The bar graph shows the distribution of riRNAs among Alu subfamilies. (b) Northern blot detected riRNAs in a time-course study. miR106a and U6 were used as controls. (c) Biotin-labeling of AGO3-immunoprecipitated RNAs from control and RA-treated cells and chemiluminescence detection. Due to the biotin conjugation, the putative miRNA appeared ~25nt. as pointed by the arrow. (d) AGO3-immunoprecipitated RNAs were analyzed by Northern blot (left gel), RT-PCR (middle upper gel) and RT-qPCR (mean±SEM; **p<0.01). RNA pull-down assays with biotin-conjugated riRNA oligonucleotides, both the 26nt. and 56nt. sequences, confirmed interaction with AGO3 (middle lower gel). (e) Northern blot examined the full-length DR2 Alu transcripts and riRNAs in Ntera2 transfected with two individual DICER siRNAs. siCTL: control siRNA. (f) Northern blot showed that overexpression of wild-type DICER (wtDicer), but not slicer-deficient mutant (mutDicer), in DICER knock-down cells, reduced the full length DR2 Alu transcripts and caused re-appearance of riRNAs. (g) Northern showed that among AGO1–4 and PIWIL1–4, only AGO3 knock-down reduced both riRNA (left two panels) and the full-length DR2 Alu RNA (right panel; two different AGO3 siRNAs were used). (h) RT-qPCR showed that in AGO3 knock-down Ntera2 cells, microinjected T7 DR2 Alu RNAs were rapidly decreased. Blue squares represent control siRNA and orange squares AGO3 siRNA.