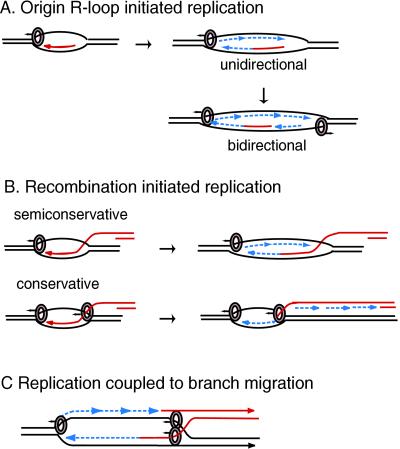
Figure 7.
Models for the role of 41 helicase in origin-initiated and recombination-initiated replication. Helicase hexamers (circles) are shown on the strands they surround. Arrows above the circles show the direction of helicase movement. Arrowheads indicate the 3′ strand ends. RNA and invading DNA molecules are in red. Newly synthesized DNA is a dashed line in blue. (A) One or two helicases are required, respectively, for unidirectional or bidirectional replication from an origin. The helicase at each fork opens the duplex ahead of the polymerase and enables primer synthesis by the primase. (B) Semiconservative and conservative replication at forks initiated by recombination. In semiconservative replication, the displaced strand serves as the lagging strand template, and the helicase role is like that at an origin. In conservative replication, the invading strand serves as the lagging strand template. The helicase at the right catalyzes the branch migration needed for reannealing the duplex behind the leading strand and also interacts with the primase on the invading strand. (C) Semiconservative replication coupled to branch migration at a four-way junction. Helicase at the fork opens the duplex and interacts with primase. Branch migration is catalyzed by 5′ to 3′ movement of helicase on the four-way junction behind the leading strand polymerase. Two helicase hexamers are shown on the junction because the products of unwinding a cruciform with nonhomologous arms are most easily explained by two 41 helicases unwinding the cruciform simultaneously (see Fig. 4). However, it remains to be determined whether a single hexamer is sufficient for strand exchange at a Holliday junction. See text for further discussion. Adapted from ref. 9.