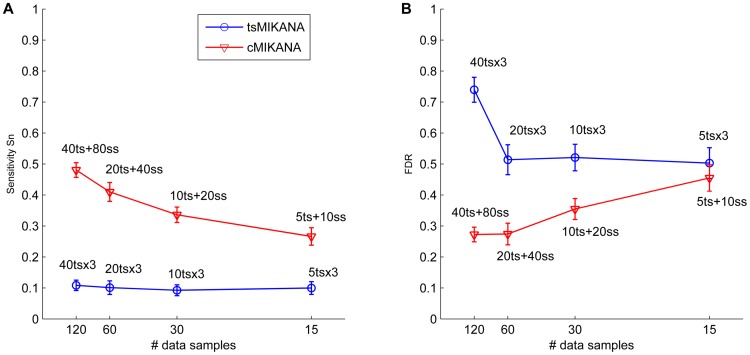
Figure 2. The performance of tsMIKANA and cMIKANA inference methods on the same size of data samples for scale-free networks with 100 genes.
The sensitivities and false discovery rates (FDRs) from MIKANA inference methods with time-series data only (tsMIKANA) and with the combination of steady-state and time-series data (cMIKANA) are compared. We reconstructed tsMIKANA network models from time-series datasets – each contained 5, 10, 20 and 40 data samples in 3 replicates, providing 15, 30, 60 and 120 data samples, respectively. We also reconstructed cMIKANA network models from the combined datasets with the same size – containing time-series data sample from 1 temporal experiment (5, 10, 20 and 40 time-series data samples, respectively) and the remainder samples were collected from several knockdown experiments (containing 10, 20, 40 and 80 steady-state data samples, respectively). 10% noise was added to both time-series and steady-state data. Being reconstructed from the same sizes of data, cMIKANA models showed higher sensitivity and relatively lower FDRs compared with tsMIKANA models, suggesting the prediction of gene regulatory interactions could be improved by incorporating steady-state data.