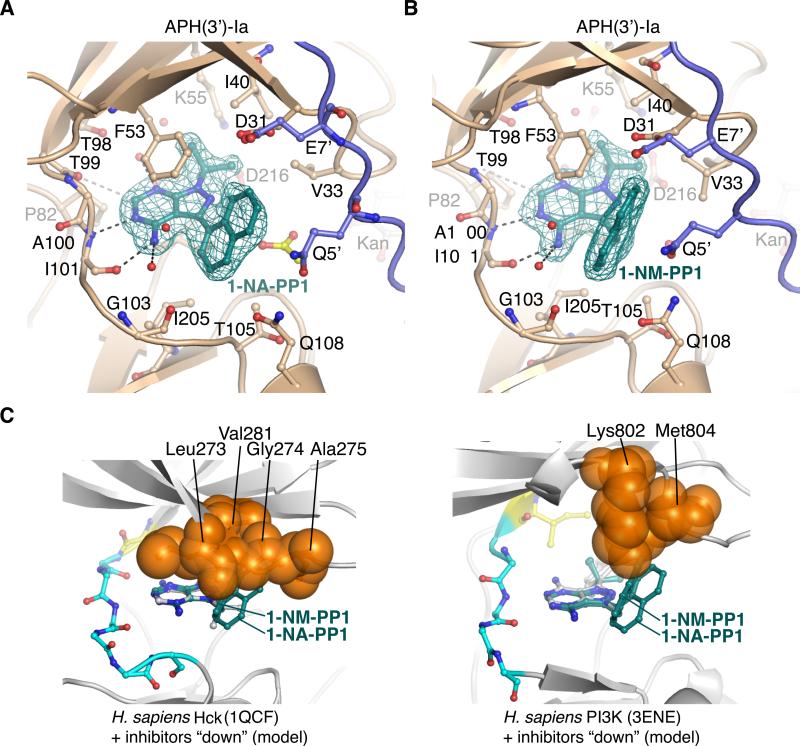
Figure 6. Structural analysis of C3-substituted compounds 1-NA-PP1 and 1-NM-PP1 bound to APH(3’)-Ia and modeling into ePKs.
A) Structure of APH(3’)-Ia•kanamycin•1-NA-PP1 complex. B) Structure of APH(3’)-Ia•kanamycin•1-NM-PP1 complex. Electron density shown for both inhibitors is simulated annealing Fo-Fc density at 3.0 σ. Symmetry-related protein chains contributing to interactions with the inhibitors are shown in purple. Acetate molecule shown in yellow. C) Modeling of 1-NA-PP1 and 1-NM-PP1 in “down” orientation into ATP-binding sites of H. sapiens Hck (PDB accession 1QCF, left) and PI3K (PDB accession 3ENE, right). Experimental structures of PP1 from the 1QCF and 3ENE structures are shown in grey sticks. Enzyme residues from ePKs expected to clash with 1-NA-PP1 and 1-NM-PP1 are shown as orange spheres.