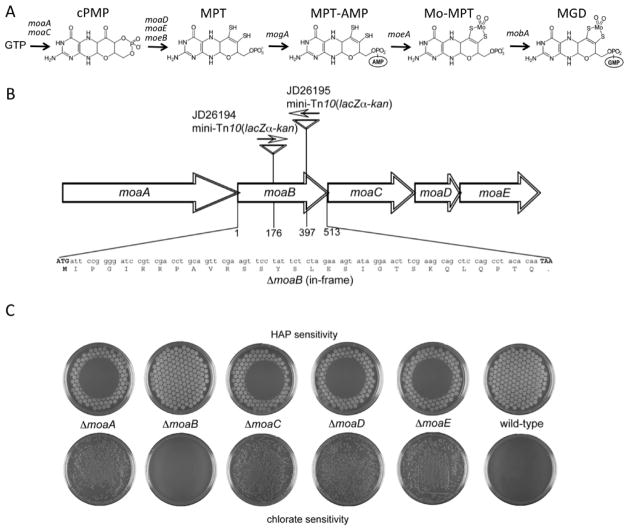
Fig. 1.
Effect of in-frame deletions of the individual genes of the E. coli moaABCDE operon on Moco-dependent activities. A. Steps in Moco biosynthesis and the responsible genes (Schwarz et al., 2009). B. Organization of the moaABCDE operon. Indicated are the locations of the moaB::mini-Tn10(lacZα-kan) insertions as present in strains JD26194 and JD26195, as well as the endpoints of the moaB deletion showing the ATG start and TAA stop codon as well as the remaining in-frame “scar” of 28 residues (see Materials and methods). All other moa genes were deleted in identical manner. C. HAP- or chlorate-sensitivity of the in-frame deletion mutants (NR17378 - NR17382) and the wild-type strain (NR10836). For the HAP-sensitivity test, cells were transferred using a multi-prong replicator to a minimal-medium plate, and 50 μg of HAP was applied onto the center of the plate. Plates were incubated overnight at 37 °C. For the chlorate-sensitivity test, cells were plated on LB plates containing 5 mM potassium chlorate and incubated 48 h at 37 °C under anaerobic conditions.