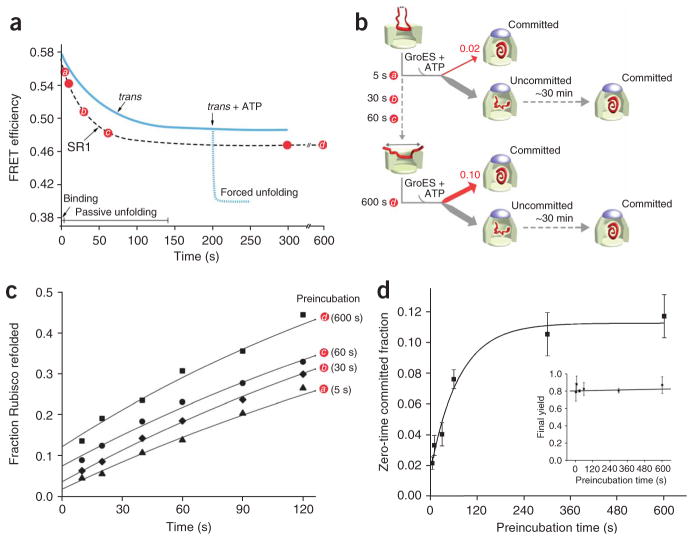
Figure 7.
The fraction of non-native Rubisco that commits to the native state in a single round of GroES encapsulation increases in proportion to the extent of unfolding. (a) The slow, passive unfolding of Rubisco on SR1 can be used to examine the linkage between productive folding and substrate-protein unfolding. The progress curve of passive Rubisco unfolding by SR1, monitored by FRET at 25 °C, is shown schematically. For comparison, a schematic of the progress curve of Rubisco unfolding on the trans ring of an ADP bullet, and the extent of forced unfolding driven by ATP binding to the trans ring, are also shown. (b) Schematic of the experiment. Denatured Rubisco bound to excess SR1 was incubated for different preincubation times at 25 °C (indicated by red circles and letters a–d) to allow passive unfolding before the addition of GroES and ATP. Because SR1 shows a dramatically reduced capacity to support forced unfolding upon ATP binding (Fig. 6), the preincubation time primarily sets the level of unfolding imposed on the bound Rubisco monomer. Following addition of ATP and GroES, the encapsulated Rubisco is allowed to fold within the enclosed SR1–GroES cavity. (c) Rubisco folding is monitored as a function of time following GroES and ATP addition for several different preincubation times. Although the overall refolding kinetics are similar, the fraction of Rubisco that commits to the native state in the first few seconds following ATP and GroES binding increases considerably at longer preincubation times. (d) The extent to which unfolding on SR1 enhances the initial commitment of Rubisco to the native state is quantified by extrapolating refolding curves like those in c to zero preincubation time for a series of identical experiments (n = 3; error bars show one s.d.). The inset in d shows the final native Rubisco yield (following 30 min of folding within the SR1–GroES complex) observed at each preincubation point.