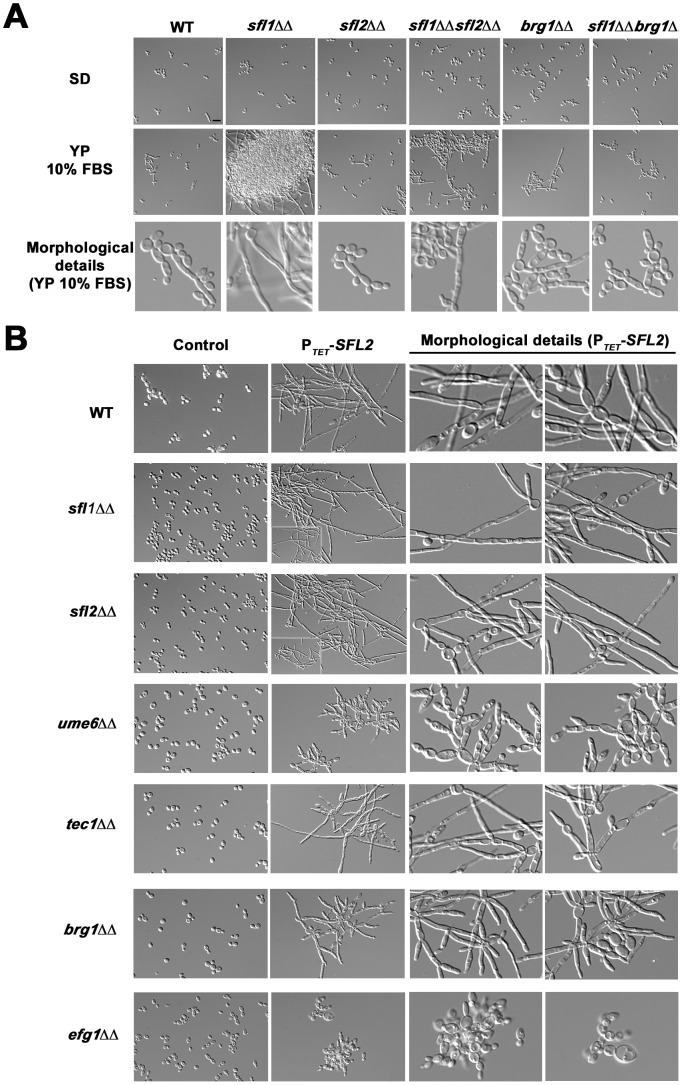
Figure 7. Genetic interactions of SFL1 and SFL2 with their transcriptional target genes encoding key regulators of hyphal development.
(A) The wild-type SC5314 (WT) together with the homozygous sfl1 (sfl1ΔΔ, CEC2001), sfl2 (sfl2ΔΔ,CEC1535), brg1 (brg1ΔΔ, CEC2058), the double homozygous sfl1, sfl2 (sfl1ΔΔ sfl2ΔΔ, CEC2658) and sfl1, brg1 (sfl1ΔΔ brg1ΔΔ, CEC2840) mutants were grown in yeast-promoting (SD at 30°C for 6 h30 min) or sub-hypha-inducing (YP 10% FBS at 30°C for 6 h30 min) conditions and observed microscopically. Scale bar = 10 µm. The detailed cell morphology of each strain grown in YP 10% FBS are shown (Morphological details, bottom panel) (B) The pNIMX expression system [41] was used to drive anhydrotetracycline-dependent overexpression of SFL2 (PTET-SFL2) in a wild-type (WT, BWP17AH complemented for uracil auxotrophy) or in different homozygous mutant backgrounds, including sfl1Δ/sfl1Δ (sfl1ΔΔ), sfl2Δ/sfl2Δ (sfl2ΔΔ), ume6Δ/ume6Δ (ume6ΔΔ), tec1Δ/tec1Δ (tec1ΔΔ), brg1Δ/brg1Δ (brg1ΔΔ) and efg1Δ/efg1Δ (efg1ΔΔ) (Table 1). All strains were grown in YPD medium at 30°C during 18 hours in the presence of 3 µg/ml of anhydrotetracycline before microscopic examination. As a control, the same growth conditions were also used with all strain backgrounds carrying the empty plasmid (CIp10, Control). Two different fields with detailed cell morphology of each strain overexpressing SFL2 are shown (Morphological details, right panels).