Figure 10. Model of Sfl1p and Sfl2p regulatory network.
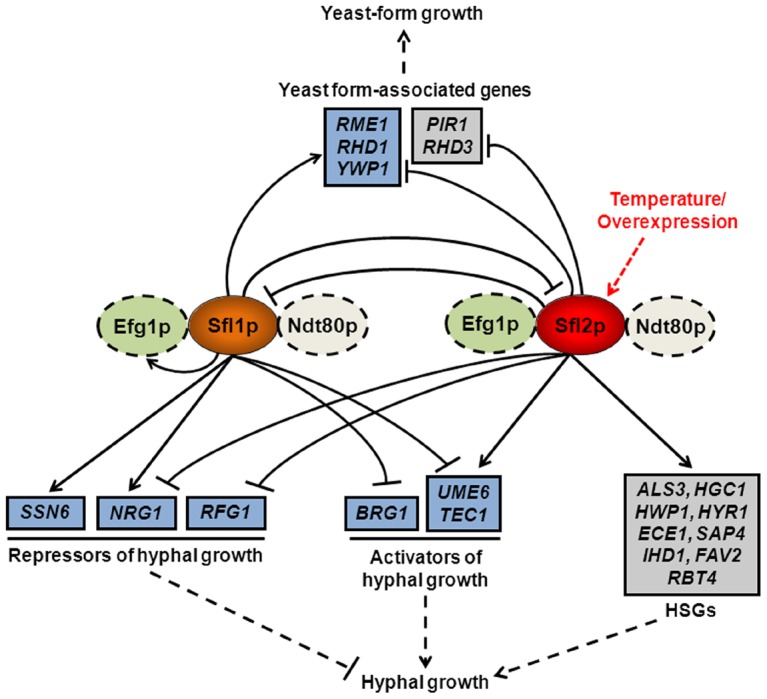
Sfl2p (red oval), which induces hyphal growth in response to temperature increase or upon overexpression (red dashed arrow), and Sfl1p (orange oval) bind directly, together with Efg1p and Ndt80p depending on growth conditions (green and white ovals, respectively; dashed lines indicate hypothetical physical and/or functional interaction), to the promoter of common (blue boxes) target genes encoding major transcriptional activators (UME6, TEC1 and BRG1) or repressors (NRG1, RFG1, SSN6) of hyphal growth as well as to the promoter of genes associated with yeast-form growth (RME1, RHD1 and YWP1) and modulate the expression of many of them (for simplicity, only modulatory direct interactions are shown i.e. both binding at and transcriptional modulation of a given target; arrowed lines indicate direct upregulation whereas blunt lines indicate direct downregulation). On the other hand, Sfl2p directly upregulates the expression of specific targets (grey boxes), including a high proportion of hyphal-specific genes (HSGs), while exerting a direct negative regulation on the expression of yeast-form associated genes (PIR1 and RHD3). Sfl1p and Sfl2p also exert a direct negative regulation on the expression of each other. The execution of Sfl1p or Sfl2p transcriptional control inputs allows to regulate the commitment (dashed line; blunt, inhibition; arrowed, activation) of C. albicans to form hyphae or yeast-form cells.
