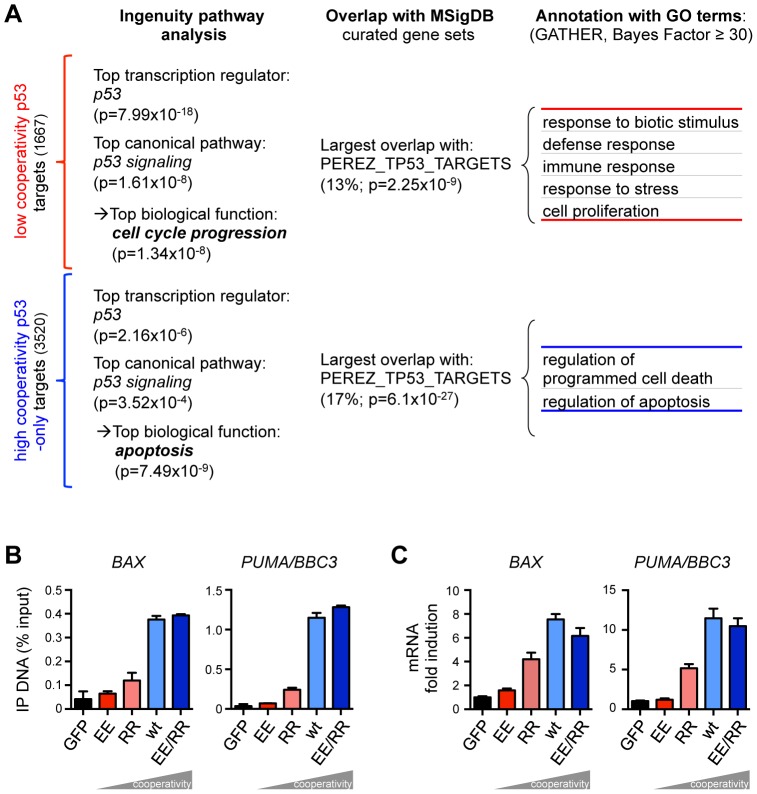
Figure 3. The requirement for DNA binding cooperativity functionally separates the p53 cistrome.
(A) Functional annotation of the neighboring genes closest to the p53 binding sites by Ingenuity Pathway Analysis or MSigDB (the percentage denotes the proportion of genes that overlap). The overlap with the MSigDB gene set PEREZ_TP53_TARGETS was annotated with Gene Ontology (GO). Shown are the GO terms unique for either list. (B) Quantification of p53 binding to the BAX and PUMA/BBC3 genes by ChIP-qPCR. Shown is the mean (±SD, n = 3) binding expressed in % of input chromatin. (C) RTqPCR quantification of BAX and PUMA/BBC3 mRNA following expression of the indicated p53 variants. Shown is the mean (±SD, n = 3) mRNA expression normalized to GAPDH and the mock sample.