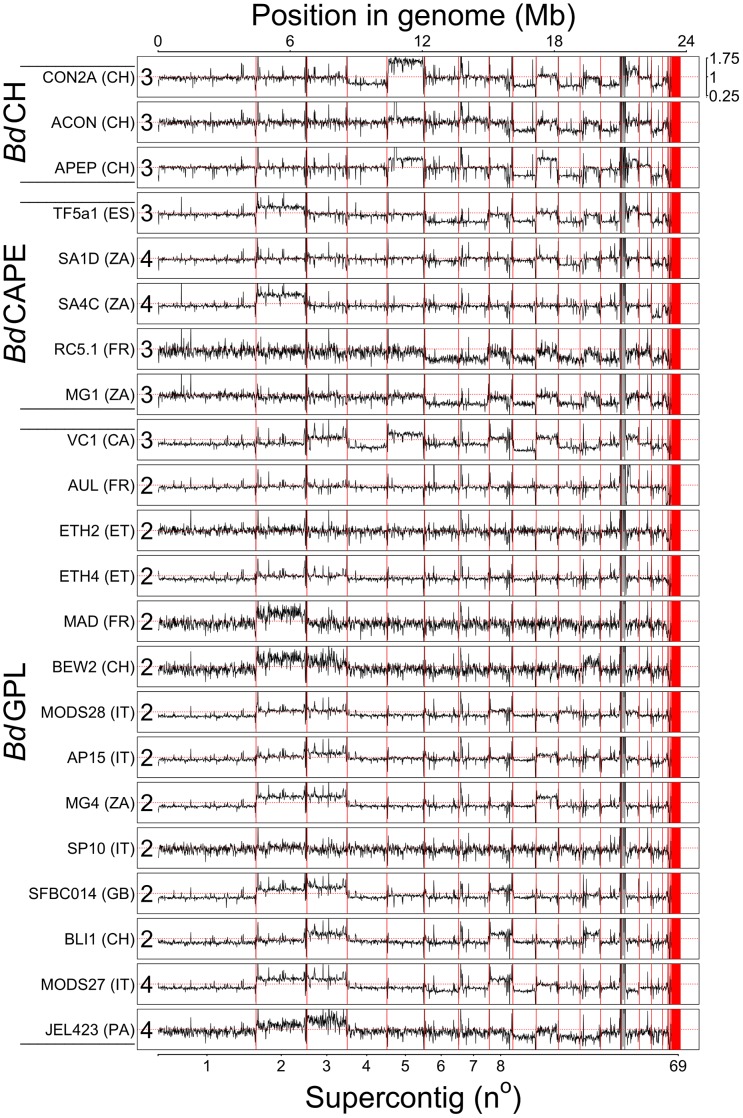
Figure 1. Read depth across 22 genomes was normalised by total alignment depth and plotted against location in the genome using a 10 Kb long non-overlapping sliding window.
Base ploidy levels were determined using allele frequencies for supercontig 1 and shown at the start of each plot. Intra-chromosome read depth is largely consistent amongst the isolates, except over supercontig 14 due to a long stretch of rDNA. Shifts in read-depth between chromosomes demonstrate variation in chromosome copy number.