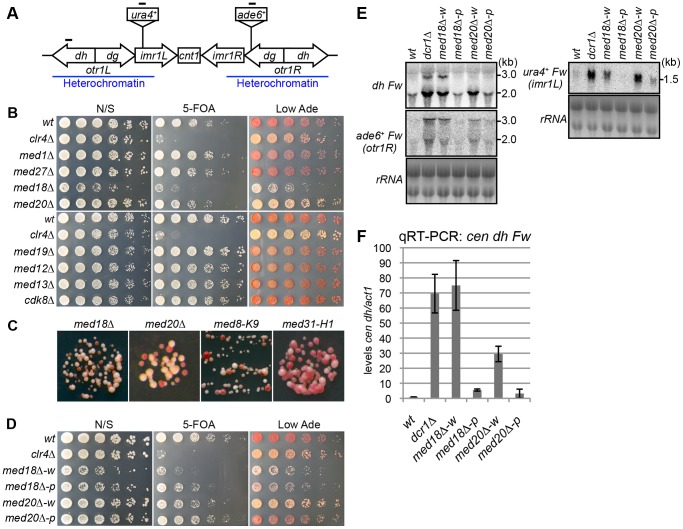
Figure 1. Mediator is required for heterochromatic silencing at the pericentromere.
(A) Schematic of fission yeast centromere 1. Locations of ura4 and ade6 reporter genes inserted within the pericentromeric region are shown (imr1L::ura4+ and otr1R::ade6). Black bars indicate the location of primers or probes used for ChIP, RT-PCR and northern analysis. (B) Silencing assay at the pericentromere. Shown are the results of serial dilutions of the indicated strains spotted onto non-selective media (N/S), medium with 5-fluoroorotic acid (5-FOA), and medium with a limited amount of adenine (Low Ade) to assay ura4+ and ade6+ expression. (C) Spots on Low Ade medium using Mediator mutants (med18Δ, med20Δ, med8-K9 and med31-H1), which are defective in heterochromatic silencing at the pericentromere. (D) Silencing assay at the pericentromere. Shown are the results of serial dilutions of the indicated strains spotted onto N/S, 5-FOA and Low Ade media to assay ura4+ and ade6+ expression. w indicates white epiclones (med18Δ-w and med20Δ-w), and p indicates pink epiclones (med18Δ-p and med20Δ-p). (E) Northern Analysis of dh, otr1R::ade6 and imr1L::ura4 forward strand transcripts in wild-type (wt) and mutant cells using oligonucleotide probes. rRNA was used as a loading control. (F) Quantitative RT-PCR analysis of cen dh forward transcript levels relative to a control act1+, normalized to the wild type in the indicated strains. Error bars show the standard error of the mean (n = 3).