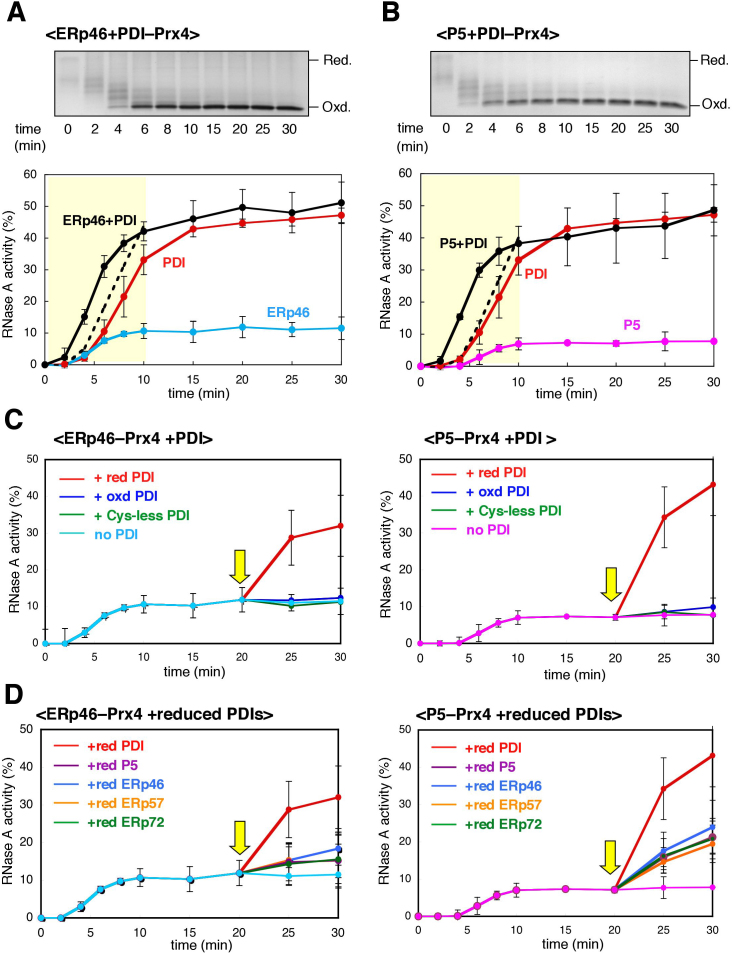
Figure 6. Synergistic action of PDIs during Prx4-driven oxidative folding.
(A) Effects of PDI on oxidative folding of RNase A catalyzed by Prx4 and ERp46. RNase A was refolded for the indicated times by Prx4 and ERp46 in the presence of an amount of PDI equimolar to that of ERp46 (2 μM) in buffer containing H2O2. The reaction was quenched by cysteine alkylation with AMS, and the products were subjected to non-reducing SDS-PAGE (upper panel). The representative non-reducing gel images of the three independent experiments are shown. The gel data is cropped to highlight the time course of RNase A oxidation. Time course of recovery of RNase A activity was measured by the change in Abs295, using cCMP as a substrate (lower panel). Values represent the mean ± SD from three independent experiments. The dotted lines indicate the result of the simple addition of the recovery curves for Prx4-PDI and Prx4-ERp46 for the refolding time 0–10 min. (B) Effects of PDI on oxidative folding of RNase A catalyzed by Prx4 and P5. Procedures were as described in (A), with the exception that P5 replaced ERp46 in the reaction mixture. The representative non-reducing gel images of the three independent experiments are shown. The gel data is cropped to highlight the time course of RNase A oxidation. (C) Recovery of RNase A activity by the action of PDI. Reduced and denatured RNaseA was pre-incubated with Prx4 and either ERp46 (left) or P5 (right) in buffer containing H2O2 for 20 min, followed by the addition of reduced, oxidized, or Cys-less PDI (5 μM) (yellow arrow). Values are the mean ± SD of three independent experiments. (D) Effects of reduced PDIs on recovery of RNase A activity. Procedures were as described in (C), with the exception that, after the 20-min pre-incubation, the reduced forms of the indicated PDIs (5 μM) were added to the reaction solution (yellow arrow). Values are the mean ± SD of three independent experiments.