Table 3.
CNVs detected by MATCHCLIP in 20 exome sequenced samples, including 10 samples with long axial length (Long AL) and 10 samples with short axial length (Short AL).
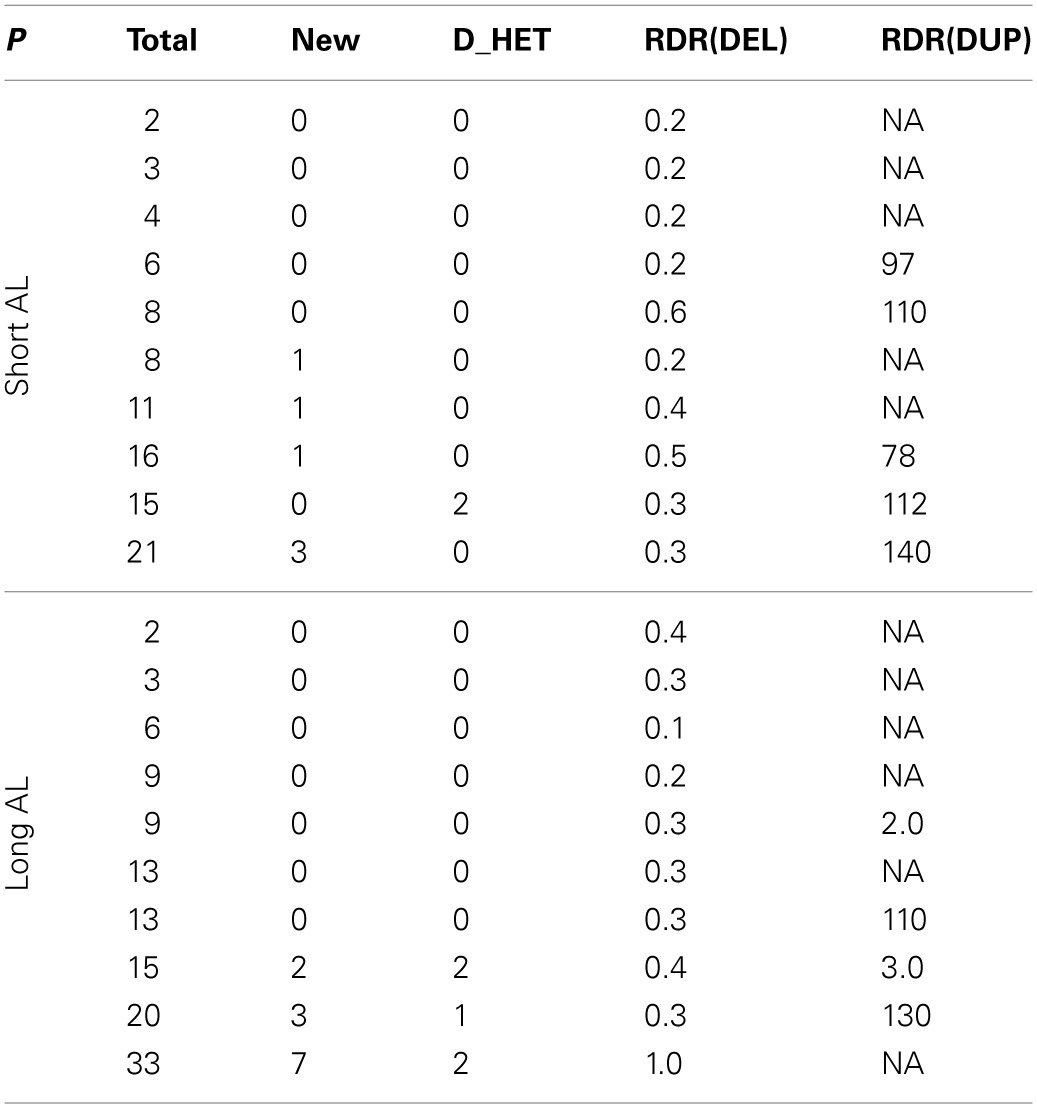
Total, number of CNVs longer than 500 basepairs; New, number of CNVs that do not overlap with any in the estd59 database (1000 Genomes Project Consortium, 2010); D_HET, number of deletion CNVs that has heterozygous sites in deleted region, where ygosity was called using samtools' mpileup function and bcftools; RDR(DEL/DUP), averaged read depth ratios (RDRs) of the read depth inside a CNV region to the read depth outside a CNV region. The outer regions include 3000 bases before and 3000 after the CNV region. NA represents no duplications were detected.
