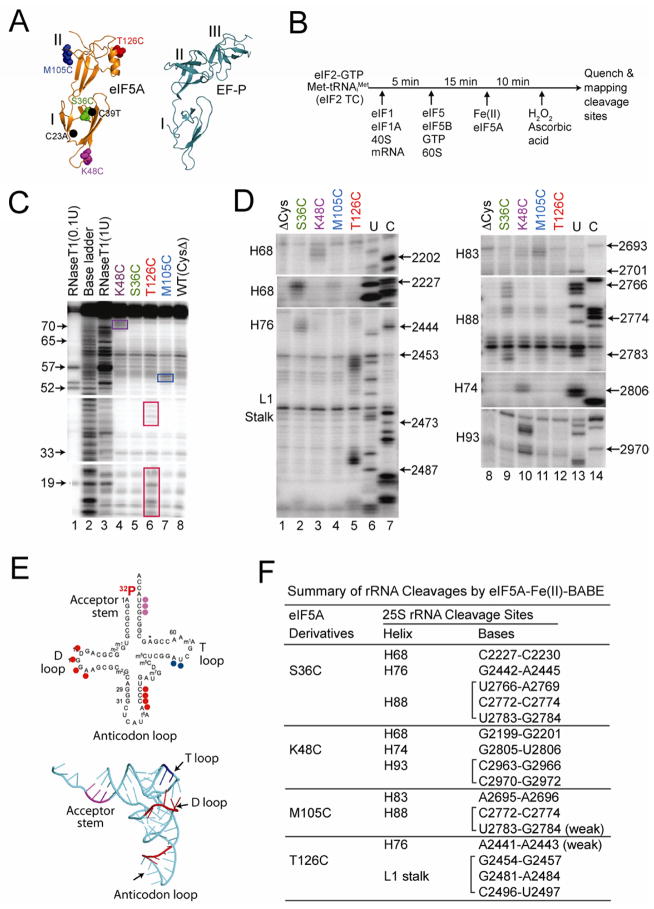
Figure 6. Directed Hydroxyl Radical Probing of eIF5A Binding to 80S Ribosomal Complexes.
(A) Ribbon representation of T. thermophilus EFP (right panel, pdb 3HUW, (Blaha et al., 2009)) and yeast eIF5A (left panel, pdb 3ER0) showing the protein domains (Roman numerals), the positions of the C23A and C39T mutations (black dots) that removed the native Cys residues in eIF5A, and the sites (Spheres representation) of Cys mutations for tethering Fe(II): Ser36 (green), Lys48 (magenta), Met105 (blue) and Thr126 (red).
(B) Scheme for directed hydroxyl radical cleavage by Fe(II)-BABE modified forms of eIF5A in 80S complexes.
(C) Directed hydroxyl radical cleavage of Met-[32P]tRNAiMet by Fe(II)-BABE- derivatized eIF5A in 80S complexes. Cleavage products were resolved on 10% (w/v) denatured polyacrylamide gels, and cleavage sites on [32P]tRNAiMet were determined by comparison to samples containing eIF5A-ΔC [WT(CysΔ), lane 8]. The tRNA ladders were prepared by digesting Met-[32P]tRNAiMet with RNase T1 (cleaves 3′ of G residue) or by base cleavage (lane 2). The tRNA residue numbers are shown at the left, and cleavage fragments are boxed.
(D) Primer extension analysis of 25S rRNA cleavage fragments produced by Fe(II)-tethered to the indicated positions in eIF5A. U, C: 25S rRNA sequencing reactions using reverse transcriptase and dideoxynucleotides ddATP and ddGTP, respectively. 25S rRNA helices and the position of the L1 stalk are indicated on the left.
(E) Sites of eIF5A-Fe(II)-BABE cleavages are shown on the secondary (left) and three- dimensional (pdb 1YFG, (Basavappa and Sigler, 1991)) structures of tRNAiMet. Cleavage sites are color-coded according to the site where Fe(II) was tethered on eIF5A (see panel A).
(F) Summary of 25S rRNA cleavages by eIF5A-Fe(II)-BABE derivatives.