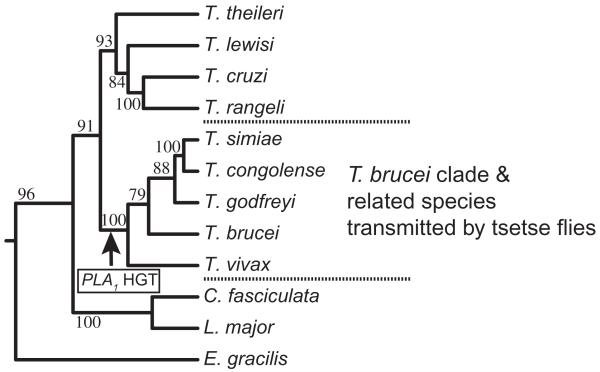
Fig. 8.
Proposed HGT point for S. glossinidius PLA1. The arrow indicates the proposed acquisition of S. glossinidius PLA1 by an ancestor of T. brucei. The maximum likelihood tree was calculated using the Phylip v3.6 Dnamlk algorithm and based on a MUSCLE alignment of glycosomal glyceraldehyde 3-phosphate dehydrogenase (gGAPDH) genes from various kinetoplastids. Values at nodes are maximum likelihood bootstrap values (%) obtained from 100 replicates (Seqboot). The tree is rooted to the corresponding gene of Euglena gracilis. The tree phylogenies agree very well with the more detailed and exact gGAPDH trees previously published (Hamilton et al., 2004; 2005). The gGAPDH sequences were obtained from GenBank and their accession numbers are: Trypanosoma theileri, AJ620282; Trypanosoma lewisi, AJ620272; Trypanosoma cruzi, AJ620269; Trypanosoma rangeli, AF053742; Trypanosoma simiae, AJ620293; Trypanosoma congolense, AJ620291; Trypanosoma godfreyi, AJ620292; Trypanosoma brucei, AJ620284; Trypanosoma vivax, AJ620295; Crithidia fasciculata, AF047493; Leishmania major, AF047497; Euglena gracilis, L21903.