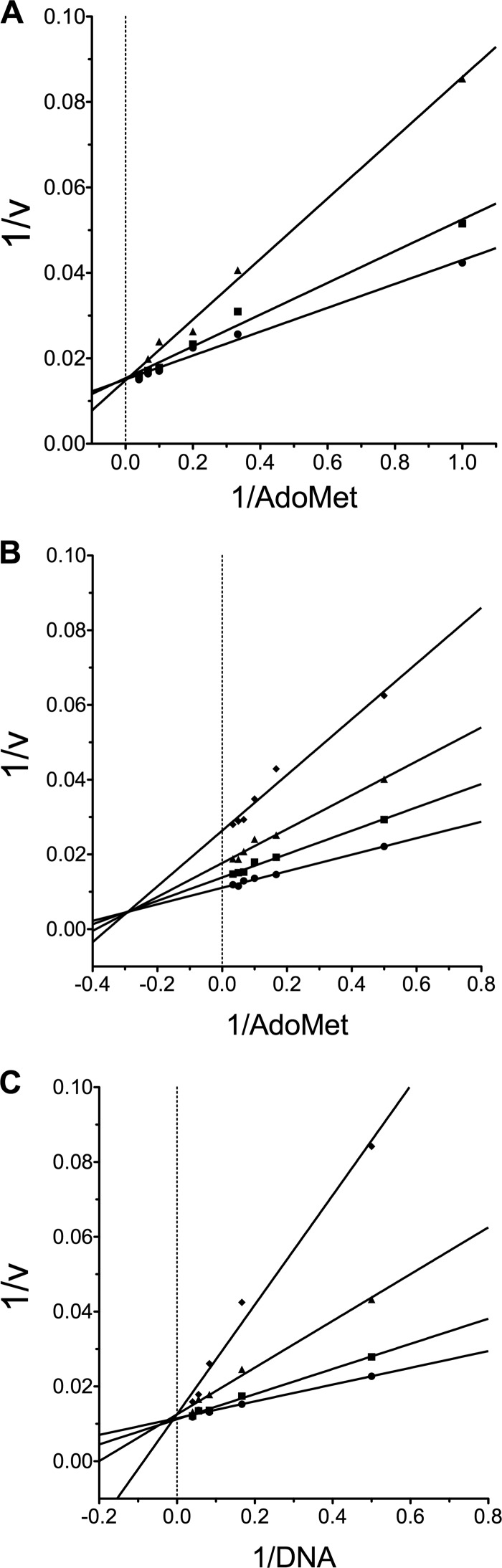
FIGURE 4.
LCA is a DNA-competitive inhibitor. A, shown are AdoMet-dependent SGI-1027 inhibition kinetics. SGI-1027 was used as an inhibitor in reactions containing 20 nm DNA (∼13× Km,DNA), and varying concentrations of AdoMet (1–25 μm) and initial velocities (relative fluorescence units/min) were determined. The double reciprocal plot of the data without SGI-1027 (●), 0.5 μm SGI-1027 (■), and 2 μm SGI-1027 (▴) shows that SGI-1027 is competitive with respect to AdoMet. Using nonlinear regression to fit the alteration in Kmapparent, a Ki of 1.5 ± 0.3 μm was determined. B, shown are AdoMet-dependent LCA inhibition kinetics. LCA was used as an inhibitor in reactions containing 20 nm DNA and varying concentrations of AdoMet (2–30 μm). The double reciprocal plot of the data without LCA (●), 0.4 μm LCA (■), 0.8 μm LCA (▴), and 1.5 μm LCA (♦) shows mixed inhibition, indicating that LCA binds both the free enzyme and the enzyme-AdoMet complex. Using nonlinear regression to fit the data (supplemental Fig. S5), a Ki,a of 640 ± 200 nm and a KI,b of 1.4 ± 0.4 μm was determined. C, DNA-dependent LCA inhibition kinetics are shown. Here, LCA was used as an inhibitor in reactions containing 30 μm AdoMet (∼15× Km,AdoMet) and varying concentrations of DNA (2–25 nm). The double reciprocal plot of the data without LCA (●), 0.4 μm LCA (■), 0.8 μm LCA (▴), and 1.5 μm LCA (♦) is indicative of competitive inhibition. Using nonlinear regression to fit the data (supplemental Fig. S5) gives a Ki of 310 ± 80 nm. In all cases, 1 nm RFTS-lacking Dnmt1 (621–1600) was used.