Background: The PTPN23 gene is a candidate tumor suppressor involved in the tumorigenesis of various organs.
Results: Expression of PTPN23 in testicular germ cell tumor cells is negatively regulated by miR-142-3p.
Conclusion: A lack of PTPN23 protein expression in human TGCTs is inversely correlated with miR-142-3p expression.
Significance: Loss of PTPN23 expression mediated by miR-142--3p may be a key event in the pathogenesis of TGCTs.
Keywords: MicroRNA, Testis, Tumor, Tumor Cell Biology, Tumor Suppressor Gene, Testicular Germ Cell Tumor
Abstract
Protein-tyrosine phosphatase non-receptor type 23 (PTPN23) is a candidate tumor suppressor involved in the tumorigenesis of various organs. However, its physiological role(s) and detailed expression profile(s) have not yet been elucidated. We investigated the function and regulation of PTPN23 in the formation of testicular germ cell tumors (TGCTs). Expression of PTPN23 in human TGCT cell lines was significantly lower than that in spermatogonial stem cells in mice. Overexpression of PTPN23 in NEC8, a human TGCT cell line, suppressed soft agar colony formation in vitro and tumor formation in nude mice in vivo. These data indicate that PTPN23 functions as a tumor suppressor in TGCTs. Multiple computational algorithms predicted that the 3′ UTR of human PTPN23 is a target for miR-142-3p. A luciferase reporter assay confirmed that miR-142-3p bound directly to the 3′ UTR of PTPN23. Introduction of pre-miR-142 in the PTPN23 transfectant of NEC8 led to suppressed expression of PTPN23 and increased soft agar colony formation. Quantitative RT-PCR data revealed a significantly higher expression of miR-142-3p in human seminomas compared with normal testes. No difference in mRNA expression between seminoma and non-seminoma samples was detected by in situ hybridization. Both quantitative RT-PCR and immunohistochemical analyses revealed that PTPN23 expression was significantly lower in TGCTs than in normal testicular tissues. Finally, a lack of PTPN23 protein expression in human TGCTs correlated with a relatively higher miR-142-3p expression. These data suggest that PTPN23 is a tumor suppressor and that repression of PTPN23 expression by miR-142-3p plays an important role in the pathogenesis of TGCTs.
Introduction
Phosphorylation of tyrosine residues plays a critical role in the regulation of cellular functions and is a central feature of the signaling cascades involved in oncogenesis. The regulation of tyrosine phosphorylation is coordinated by protein-tyrosine kinases and phosphatases (PTPs)2. Perturbation of protein-tyrosine kinase signaling by overexpression or by mutations that result in deregulated kinase activity is often observed in cases of malignant transformation.
On the other hand, PTPs are predicted to have tumor-suppressive functions because of their antagonism of kinase activity. We have found previously that the Ptpn23 gene is expressed in male germ line stem cells (1). PTPN23 (or His domain-containing protein-tyrosine phosphatase) belongs to the non-receptor class subfamily of the PTP family, several members of which have been implicated in tumor suppression (2). For example, loss of PTPN13 in non-small-cell lung cancer was shown to be associated with increased signaling through the epidermal growth factor receptor and HER2 tyrosine kinase receptors (3).
PTPN23 encodes a 1636-amino acid protein, the most striking feature of which is the sequence at the PTP active center (VHCSSG), which is distinct from the invariant sequence present in PTPs identified previously (VHCSAG). The gene encodes a BRO1-like protein (which plays a role in endosomal targeting), a histidine-rich domain, a PTP-like domain, and a protein-destabilizing sequence (PEST motif) (4). PTPN23 is highly evolutionarily conserved from yeast to human, and the Ptpn23 homozygous deletion mouse is embryonic lethal at around embryonic day 9.5, suggesting that PTPN23 is essential during the early stages of development (5).
Cao et al. (6) showed that PTP-TD14, the rat homolog of PTPN23, inhibits activated H-ras-mediated transformation of NIH-3T3 cells. Later, a hemizygous missense mutation within the histidine-rich domain in the human PTPN23 gene was identified in a small-cell lung cancer cell line (4). Several functions of PTPN23 have been reported since then, including its role in the regulation of endothelial cell motility by modulating tyrosine phosphorylation of focal adhesion kinase (FAK) (7) and its interaction with SRC (8). Furthermore, expression of PTPN23 reduced the colony-forming capacity of human renal cancer cells, a process independent of catalytic protein-tyrosine phosphatase activity (9). In addition, a functional genomic screening using RNA interference identified PTPN23 as a gene involved in controlling ciliogenesis (10). Functional assays showed that silencing of PTPN23 markedly reduced the number of ciliated cells. Another functional screening using RNA interference showed that PTPN23 acts as a negative regulator of SRC in breast cancer to modulate cell motility and invasion (11). Very recently, Casiglioni et al. (12) showed that PTPN23 is degraded by calpain in bladder carcinoma T24 cells, and they proposed that degradation of PTPN23 might enhance cell migration and invasion.
TGCTs are the most common malignancies in adolescent and adult males aged 14–40 years. TGCTs are a heterogeneous group of neoplasms classified as seminomas or non-seminomas (embryonal carcinomas, teratomas, choriocarcinomas, and yolk sac tumors). An isochromosome of the short arm of chromosome 12 is the most common and characteristic cytogenetic aberration in TGCTs.
In addition, molecular genetic changes in human TGCTs showed 3p allele loss, suggesting the presence of a tumor suppressor gene within this region (13–15). Notably, the PTPN23 gene is located within this region (chromosome 3p21.3) in an area of the genome frequently lost in breast (16), lung (17), nasopharyngeal (18), cervical (19), and kidney (20) carcinomas. However, deletion of the PTPN23 gene in TGCTs has not yet been reported.
MicroRNAs (miRNAs), a class of small RNA molecules that negatively regulate their mRNA targets in a sequence-specific manner, are frequently dysregulated in human cancers and can act as potent oncogenes and tumor suppressor genes. miRNA overexpression has been observed in various human tumors, and these molecules target important tumor suppressors. For example, miR-21, miR-17–92, miR-221, and miR-222 target phosphatase and tensin homolog (PTEN) (21–23), and miR-372 and miR-373 target LATS2 (24).
In this study, we show that the colony-forming capacity in soft agar and tumorigenicity of a human TGCT cell line are suppressed by overexpression of PTPN23. These data indicate that PTPN23 functions as a tumor suppressor in TGCTs. Furthermore, we found that miR-142-3p bound directly to the 3′ UTR of PTPN23 and that the tumor-suppressive activity of PTPN23 was decreased by overexpression of the miR-142 precursor. In human samples, PTPN23 expression was down-regulated significantly and correlated negatively with miR-142-3p expression in TGCTs.
EXPERIMENTAL PROCEDURES
Establishment of a PTPN23-overexpressing TGCT Cell Line
NEC8 and NEC14 (human embryonal carcinoma-derived TGCT cell lines), and GC-1 (a mouse spermantogonia-derived cell line) were purchased from the ATCC. For the constitutive expression of PTPN23, PTPN23 cDNA spanning exons 1–25 (total of 5107 bp, GenBank accession number NM_015466), which includes the miR-142-3p binding site, was inserted into the pMYs-ires-EGFP retrovirus vector. The construct was then transfected into a Plat-A cell line to produce a recombinant retrovirus. NEC8 cells were infected with retroviral supernatant supplemented with Polybrene (8 μg/ml). Infected enhanced GFP-positive cells were sorted using a FACSAria (BD Biosciences). Cell lines were cultured in DMEM (Sigma-Aldrich, St. Louis, MO) supplemented with 10% FBS (Equitech Bio, Kerrville, TX) in 5% CO2 at 37 °C. For cell cycle analysis, cells grown in 1% FBS-DMEM were incubated with 0.1% Nonidet P-40-PBS and propidium iodide (2 μg/ml) at room temperature for 15 min. DNA content and cell cycle phase were determined on a FACSCalibur (BD Biosciences).
Quantitative Real-time PCR
Total RNAs and miRNAs were isolated using a miRNeasy mini kit (Qiagen, Hilden, Germany) according to the protocol of the manufacturer. RT was performed at 16 °C for 30 min, 42 °C for 30 min, and 85 °C for 5 min using the TaqMan MicroRNA RT kit (Invitrogen). Quantitative real-time PCR (qRT-PCR) detection of mature miR-142-3p was performed using the TaqMan microRNA assay and Universal PCR Master Mix (Invitrogen). The reactions were performed at 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min using a LightCycler 480 real-time PCR system (Roche Applied Science).
Inhibitor of miR-142-3p (anti-hsa-miR-142-3p, 5′-UGUAGUGUUUCCUACUUUAUGGA-3′) and negative control RNA were purchased from Qiagen. Anti-hsa-miR-142-3p or control RNA was transfected into NEC8 cells at a final concentration of 50 nm using siPORT NeoFX (Invitrogen) according to the instructions of the manufacturer. Two days after transfection, total RNAs and miRNAs were extracted and reverse-transcribed into cDNA using the PrimeScript RT mix for RT-PCR (Takara, Otsu, Japan). Real-time PCR was performed with SYBR premix Ex TaqII (Takara) and the following primers: human PTPN23 (85 bp), 5′-CCGACACTGTCAGGAACCTT-3′ (forward) and 5′-CTGATGTCCTTCAGGGAAGC-3′ (reverse); human GAPDH (66 bp), 5′-ATGTTCGTCATGGGTGTGAA-3′ (forward) and 5′-GGAGGCATTGCTGATGATCT-3′ (reverse); mouse Ptpn23 (192 bp), 5′-CTGACCACTCAGAGATGAAG-3′ (forward) and 5′-TCTGTAGTGTGGAGTTCCAC-3′ (reverse); and mouse Gapdh (132 bp), 5′-AACTTTGGCATTGTGGAAGG-3′ (forward) and 5′-GGATGCAGGGATGATGTTCT-3′ (reverse).
For the overexpression experiments, pre-miR-142-3p (Invitrogen) was transfected into GC-1 cells at a final concentration of 50 nm using siPORT NeoFX. The qRT-PCR results, recorded as threshold cycle number (Ct), were normalized against an internal control (RNU6B) and expressed as fold changes.
Western blot Analysis
Protein was extracted from control/NEC8 and PTPN23/NEC8 cells using radioimmune precipitation assay lysis buffer supplemented with a proteinase inhibitor mixture (Roche Applied Science) and subjected to electrophoresis on a Super Sep 3–10% polyacrylamide gel (Wako, Osaka, Japan) in the presence of SDS under reducing conditions. Proteins transferred onto a Hybond-P PVDF membrane (GE Healthcare) overnight at 4 °C. The blots were blocked by 5% skim milk (Difco) and incubated with primary antibody. PTPN23 protein was detected with a polyclonal anti-PTPN23 antibody (1:200 dilution, catalog no. 10472-1, ProteinTech Group, Chicago, IL) followed by incubation with HRP-conjugated secondary antibody (1:1000, GE Healthcare) for 1 h at room temperature. As a loading control experiment, the membrane was reblotted with an anti-α-tubulin antibody (1:500, catalog no. T 9026, Sigma-Aldrich). Chemiluminescence detection was carried out using ECL Plus detection reagent (GE Healthcare) and a Luminescent Image Analyzer LAS-3000 (Fujifilm, Tokyo, Japan).
In Vitro Colony Assay and in Vivo Tumorigenesis Assay in Mice
A soft agar colony assay and a tumor growth assay in nude mice were performed as described previously (25). Balb/c nu/nu male mice (6 weeks old) were purchased from Nihon SLC, Hamamatsu, Japan. All mice were maintained under a 12-h light/12-h dark cycle in a pathogen-free animal facility. All experimental procedures involving mice were preapproved by the Ethical Committee for Animal Experiments at Tokyo Metropolitan Institute of Medical Science and were performed according to the Guidelines for the Proper Conduct of Animal Experiments.
Search for miRNA Target Sequences in the 3′ UTR of PTPN23
The candidate miRNA acting on the PTPN23 gene was identified on the basis of a conservative intersection of the following mRNA target prediction tools: TargetScan, miRANDA, and PicTar.
Luciferase Reporter Assay
To examine the role of miR-142-3p regulation in PTPN23 expression, a 132-bp fragment of the 3′ UTR region of human PTPN23 containing the predicted miR-142-3p site was subcloned into the XbaI restriction site of the pGL3 luciferase vector (Promega, Madison, WI). The base pair change in the predicted miRNA seed sequence (ACACTACA to CAGTGCAG) was accomplished by site-directed mutagenesis using appropriate primers. NEC8 cells were transfected with 50 nm pre-miR-142-3p or the negative control using siPORT NeoFX. After 6 h, 50 ng of firefly luciferase reporter vector containing the wild-type or mutant PTPN23 3′ UTR sequence and 20 ng of the pRL-CMV Renilla luciferase control vector (Promega) were transfected using Lipofectamine LTX (Invitrogen). Luciferase assays were performed 48 h after transfection using the Dual-Luciferase reporter assay system (Promega). Firefly luciferase activity was normalized to Renilla luciferase activity.
Construction of a miR-142 Precursor Expression Vector
The Hsa-miR-142 precursor (287 bp) was amplified by PCR using the following primers: 5′-TTGGGGGGATCTTAGGAAGC-3′ and 5′-GGAGGGGCCGTAAGGTGCTC-3′. It was then cloned into the pEF6 vector (Invitrogen) and transfected into PTPN23/NEC8 cells, followed by blasticidin selection.
Immunohistochemistry
Immunohistochemical staining was performed on formalin-fixed paraffin-embedded specimens. After dewaxing in xylene and graded ethanol, the sections were incubated in 3% H2O2 solution for 15 min to block endogenous peroxidase activity. The sections were incubated with the anti-PTPN23 primary antibody (1:100) at 4 °C overnight and processed using the DAB system (Vector, Burlingame, CA). Paraffin sections of control/NEC8-derived solid tumors were evaluated using an anti-GFP (1:200, catalog no. A-6455, Invitrogen) in combination with Alexa Fluor 488-conjugated anti-rabbit IgG secondary antibody (catalog no. 4412, Invitrogen).
In Situ Hybridization
To investigate the cell-specific distribution of miRNA in normal testes and TGCTs, in situ hybridization was performed using 5′- and 3′- digoxigenin-labeled, locked nucleic acid-modified DNA oligonucleotides complementary to the mature miRNA (Exiqon A/S, Vedbaek, Denmark). Expression of miR-142-3p in a testis cancer tissue array (TE231, US Biomax, Rockville, MD) was also examined by in situ hybridization according to the instructions of the manufacturer.
Human Patient Samples
The following formalin-fixed paraffin-embedded tissue sections were purchased from US Biomax: HuCAT381 human testis cancer (seminoma, age 45), HuFPT151 normal human testis (age 74), and a testis cancer tissue array with normal testis control tissues (TE231). Normal human testis (T2234260, age 26) and human testis cancer (T2235260-3, seminoma, age 47) were purchased from BioChain (Newark, CA). Patient specimens (S3, seminoma, age 36; S4, seminoma, age 35; S5, seminoma, age 43; and S6, seminoma, age 34) were taken at the time of surgery at Yokohama City University Medical Center (Yokohama, Japan) between 2002 and 2012. We obtained written informed consent from all patients, and the study was approved by the Ethical Committee of the Yokohama City University Medical Center and School of Medicine. Total RNAs and miRNAs from formalin-fixed paraffin-embedded tissues were extracted using a RecoverAll total nucleic acid isolation kit (Invitrogen).
Statistical Analyses
Statistical differences in the levels of mRNA expression, data in the colony assays, and reporter activities in the luciferase assays were determined using Student's t test. A p value of <0.01 was considered to be significant.
RESULTS
Establishment of a PTPN23-overexpressing TGCT Cell Line
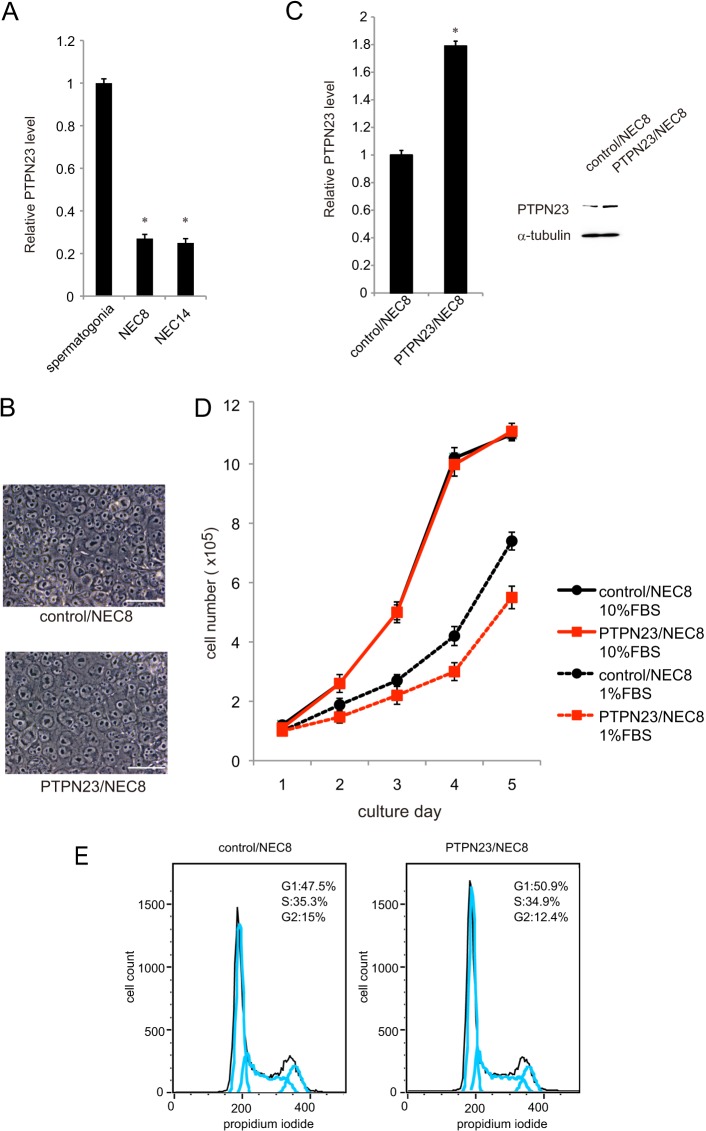
We previously identified Ptpn23 as one of the spermatogonia-specific genes. PTPN23 protein-positive cells were found near the basement membrane of the seminiferous tubules in adult mouse testes, where spermatogonial stem cells reside (Fig. 1). Because PTPN23 is a candidate tumor suppressor, we investigated the hypothesis that loss of PTPN23 function may play a role in the development of TGCTs. First, we examined PTPN23 mRNA expression in two TGCT cell lines and compared it with that in c-Kit+ spermatogonial stem cells isolated from 7-day-old mice. Notably, the levels of PTPN23 expression in both TGCT cell lines were approximately one-fourth of the Ptpn23 level in spermatogonial stem cells (Fig. 2A).
FIGURE 1.
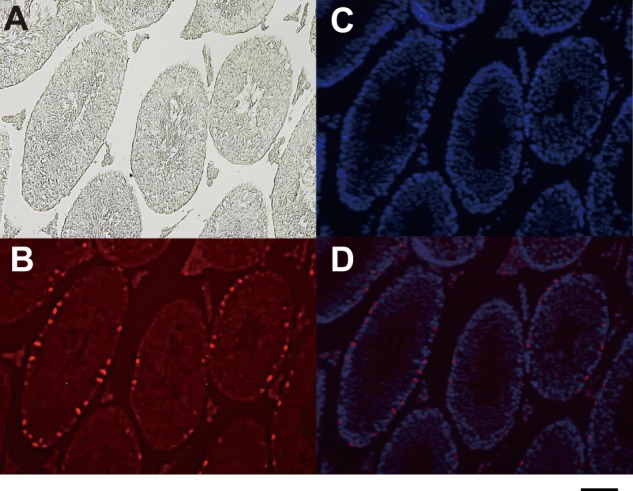
Expression of Ptpn23 protein in adult mouse testis. A, phase-contrast microphotograph of a section from adult mouse testis. B, immunofluorescent staining of the testis section with an anti-Ptpn23 antibody. C, DAPI staining of the testis section. D, merged image of B and C. Scale bar = 100 μm.
FIGURE 2.
Establishment of a PTPN23-overexpressing TGCT cell line. A, expression of Ptpn23 mRNA in c-Kit+ spermatogonial stem cells and PTPN23 mRNA in two human TGCT cell lines, NEC8 and NEC14. Expression levels were normalized against an internal control (Gapdh or GAPDH). Each value represents the mean ± S.E. (n = 3). *, p < 0.01 compared with value of c-Kit+ spermatogonial stem cells. B, morphology of NEC8 cells transfected with the control vector (control/NEC8) and NEC8 cells overexpressing PTPN23 (PTPN23/NEC8). Scale bar = 50 μm. C, left panel, expressions of PTPN23 mRNA in control/NEC8 and PTPN23/NEC8 cells. Each reaction was performed in triplicate. *, p < 0.01 compared with control/NEC8 cells. Right panel, total cell lysates from control/NEC8 and PTPN23/NEC8 cells were subjected to Western blot analysis with anti-PTPN23 or α-tubulin (control) antibodies. D, cell numbers in cultures of control/NEC8 or PTPN23/NEC8 cells in the presence of 1% or 10% FBS were counted every day. Each value represents the mean ± S.E. (n = 3). E, cell cycle profiles of control/NEC8 or PTPN23/NEC8 cells grown in 1% FBS-DMEM by FACS.
Using retroviral infection, we generated an NEC8 transfectant (PTPN23/NEC8) overexpressing PTPN23 and GFP. PTPN23/NEC8 cells were morphologically indistinguishable from control NEC8 cells (control/NEC8) (Fig. 2B), although PTPN23 expression in PTPN23/NEC8 was higher than control/NEC8 at both mRNA and protein levels (Fig. 2C). The growth rate and maximum density of PTPN23/NEC8 cells in 10%FBS-DMEM was identical to that of control/NEC8 cells (Fig. 2D). The doubling time of these cells was calculated to be 24 h from the exponential growth phase of the growth curve. However, in 1% FBS-DMEM, PTPN23/NEC8 grew more slowly than control/NEC8 cells (∼20% reduction of the growth rate) (Fig. 2D). Consistent with these data, the percentage of cells in G1 phase increased slightly under this experimental condition (Fig. 2E).
PTPN23 Overexpression in TGCT Cells Suppresses Their Anchorage-independent Growth in Vitro and Tumorigenicity in Vivo
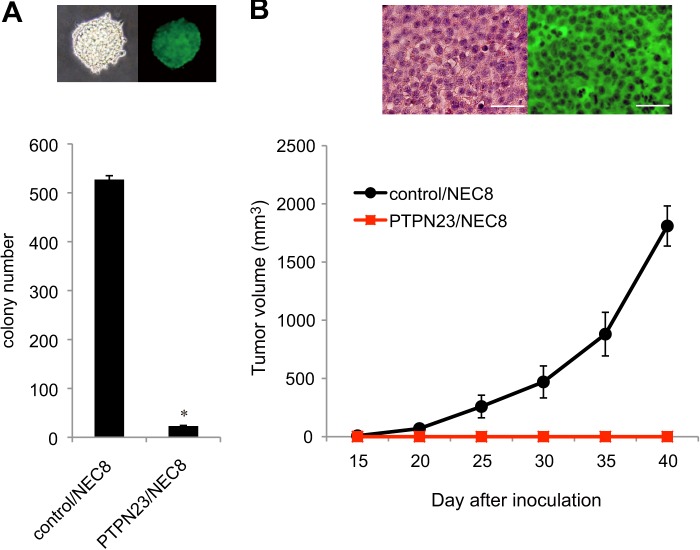
To determine whether PTPN23 has a tumor-suppressive function in TGCT cells, we performed soft agar colony formation assays and transplantation experiments in athymic nu/nu mice. The colony number of PTPN23/NEC8 cells in soft agar was only 4% of that of control/NEC8 cells (Fig. 3A). We confirmed surrogate GFP expression in the control/NEC8-derived colonies in soft agar (Fig. 3A, upper panel). These data indicate an inhibitory role of PTPN23 against the anchorage-independent growth of tumor cells.
FIGURE 3.
PTPN23 overexpression in TGCT cells suppresses anchorage-independent growth in vitro and tumorigenicity in vivo. A, control/NEC8 or PTPN23/NEC8 cells (1 × 104 cells) were inoculated into soft agar medium. After 2 weeks in culture, the number of colonies was counted. Each value represents the mean ± S.E. (n = 5) *, p < 0.01 compared with control/NEC8 cells. Top panel, bright-field image (left) and fluorescent image (right) of the control/NEC8-derived soft agar colonies. B, control/NEC8 or PTPN23/NEC8 cells (5 × 106 cells) were injected subcutaneously into athymic nude mice, and the tumor volume was measured periodically. Each value represents the mean ± S.E. (n = 3). Top panel, hematoxylin and eosin (left) and immunohistochemical staining (right) of paraffin-embedded sections of control/NEC8 cell-derived solid tumors. Scale bar = 50 μm.
Next, we examined in vivo tumorigenicity by subcutaneously inoculating control/NEC8 or PTPN23/NEC8 cells (5 × 106 cells) into nude mice. The control cells formed palpable tumors in all mice within 3 weeks of subcutaneous injection, and the tumors grew to a volume of 2000 mm3 after 6 weeks. However, none of the mice injected with PTPN23/NEC8 cells developed tumors (Fig. 3B), even 3 months after inoculation (data not shown). Taken together, these data suggest that PTPN23 is a key negative regulator for the transformed phenotypes of TGCTs. Immunohistochemical analysis of tumor xenografts excised from the mice injected with control/NEC8 cells revealed the GFP expression (Fig. 3B, upper panel), proving the donor origin of the tumors.
PTPN23 Is a Direct Target of miR-142-3p
miRNAs exert their function by regulating the expression of their downstream target gene(s). Multiple computational algorithms using TargetScan, miRANDA, and PicTar predicted that the 3′ UTR of human PTPN23 gene is a target for miR-142-3p. The 3′ UTR of PTPN23 mRNA contained a complementary nucleotide sequence for the seed region of miR-142-3p (Fig. 4A). This “seed” region of miR-142-3p was highly conserved among species, indicating its regulatory importance (Fig. 4B).
FIGURE 4.
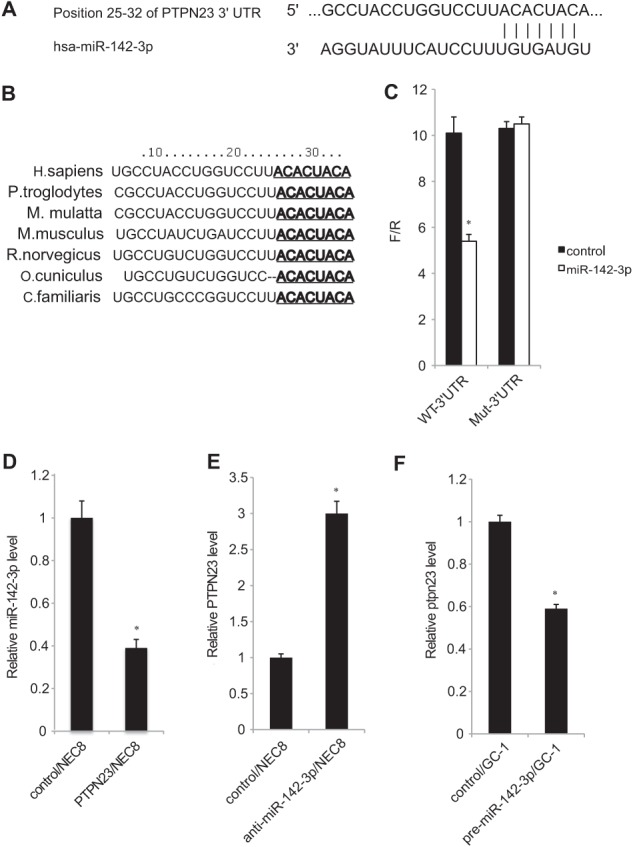
PTPN23 is a direct target of miR-142-3p. A, the miR-142-3p target site within the 3′ UTR of the human PTPN23 gene. B, comparison of the miR-142-3p seed sequence in seven different species. C, luciferase assays using reporter vectors containing the wild-type PTPN23 3′ UTR seed sequence (WT-3′ UTR) or the mutant construct in which mutations were introduced into the seed sequence (Mut-3′ UTR). NEC8 cells were transfected with the pre-miR-142-3p expression vector or the control vector in combination with one of the reporter constructs. Each value represents the mean ± S.E. (n = 3). *, p < 0.01 compared with the control. D, expression of miR-142-3p mRNA in control/NEC8 and PTPN23/NEC8 cells. Each value represents the mean ± S.E. (n = 3). *, p < 0.01 compared with control/NEC8 cells. E, NEC8 cells were transfected with anti-miR-142-3p or negative control. Total RNA was harvested 48 h after transfection and analyzed for PTPN23 mRNA expression using qRT-PCR. Each value represents the mean ± S.E. (n = 3). *, p < 0.01 compared with control/NEC8 cells. F, GC-1 cells were transfected with pre-miR-142-3p or the pre-miR-negative control. RNA was harvested 72 h after transfection and analyzed for Ptpn23 expression. Each value represents the mean ± S.E. (n = 3). *, p < 0.01 compared with control/GC-1 cells.
To experimentally verify the above possibility, the miR-142-3p binding sequence from the 3′ UTR of PTPN23 mRNA (WT-3′UTR), or its mutated version (Mut-3′UTR), was cloned downstream of the firefly luciferase reporter gene and then cotransfected into NEC8 cells along with pre-miR-142-3p or the pre-miR-negative control. The reporter activity of the luciferase gene fused with WT-3′UTR, but not Mut-3′UTR, was repressed by coexpression of miR-142-3p (Fig. 4C), demonstrating that miR-142-3p targets the 3′ UTR of PTPN23 mRNA. When we compared expression levels of miR-142-3p between PTPN23/NEC8 and control/NEC8 cells, it was decreased in the former cells (Fig. 4D). This is likely due to complex formation with the 3′UTR of exogenously expressed PTPN23 mRNA.
Next, we introduced anti-miR-142-3p into NEC8 cells and found that miR-142-3p inhibition resulted in the increase of endogenous PTPN23 mRNA (Fig. 4E). Moreover, overexpression of miR-142-3p in a mouse spermatogonia-derived cell line, GC-1, reduced the expression of Ptpn23 mRNA (Fig. 4F). Thus, we demonstrated an inverse correlation between miR-142-3p and PTPN23/Ptpn23 expressions in various male germ cell-derived cell lines.
Restoration of Tumorigenicity by Reintroduction of miR-142 in PTPN23/NEC8 Cells
To further confirm that the miR-142-3p-mediated reduction of PTPN23 restores tumorigenicity, pre-miR-142 was stably transfected into PTPN23/NEC8 cells (miR-142/PTPN23/NEC8), followed by qRT-PCR and Western blot analysis. The expression of miR-142-3p mRNA was increased greatly in miR-142/PTPN23/NEC8 cells (Fig. 5A). In these cells, PTPN23 mRNA was reduced ∼40% (Fig. 5B), and PTPN23 protein was mostly abolished (Fig. 5C) when compared with control/PTPN23/NEC8 cells. We then examined the anchorage-independent growth of miR-142/PTPN23/NEC8 cells. As expected, miR-142/PTPN23/NEC8 cells gave rise to 4 times more colonies in the soft agar culture than control/PTPN23/NEC8 cells (Fig. 5D).
FIGURE 5.
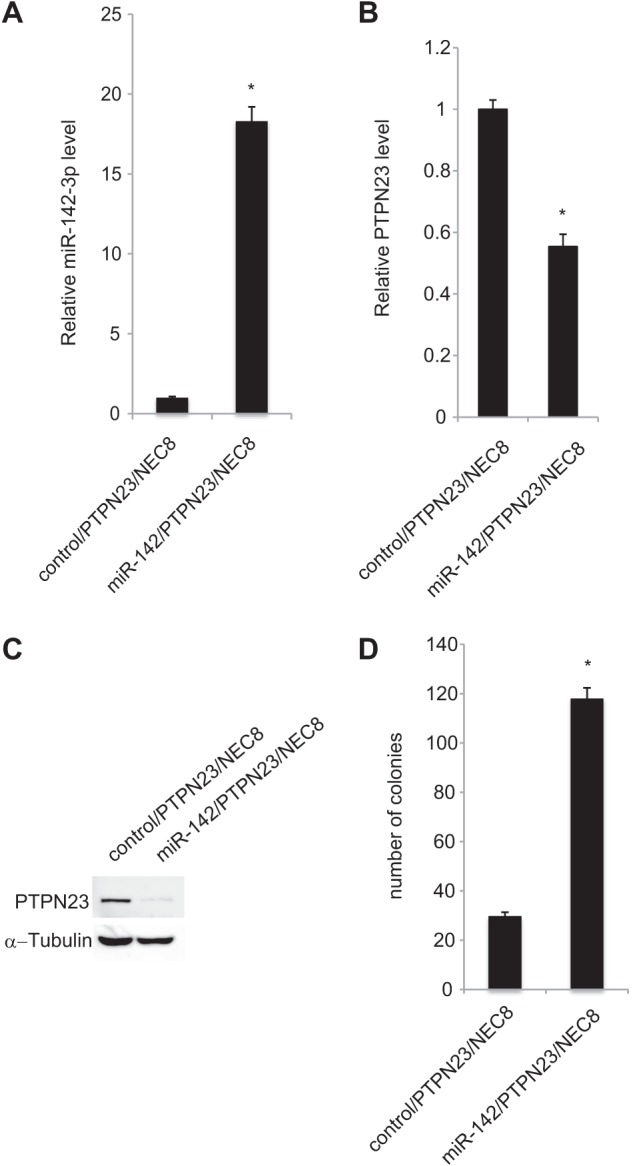
Restoration of tumorigenicity by reintroduction of miR-142 in PTPN23/NEC8. A, expression of miR-142-3p in the miR-142 precursor transfected PTPN23/NEC8 cells. Each value represents the mean ± S.E. (n = 3). *, p < 0.01 compared with control/PTPN23/NEC8. B, expression of PTPN23 in the miR-142 precursor-transfected PTPN23/NEC8 cells. Each value represents the mean ± S.E. (n = 3). *, p < 0.01 compared with control/PTPN23/NEC8. C, cell lysates from control/PTPN23/NEC8 and miR-142/PTPN23/NEC8 cells were subjected to Western blot analysis with anti-PTPN23 or α-tubulin (control) antibodies. D, restoration of the colony-forming capacity in soft agar by miR-142 transfection into PTPN23/NEC8 cells. Control/PTPN23/NEC8 or miR-142/PTPN23/NEC8 cells were inoculated into soft agar medium. After 2 weeks in culture, the number of colonies was counted. Each value represents the mean ± S.E. (n = 5). *, p < 0.01 compared with control/PTPN23/NEC8.
Negative Correlation between PTPN23 and miR-142-3p Expression in TGCTs and Normal Testes
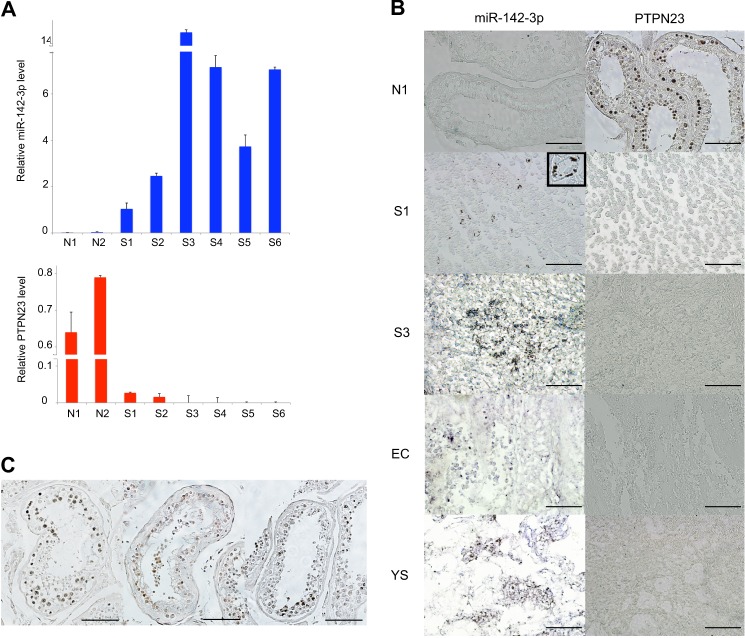
Finally, we investigated the expression of miR-142-3p and PTPN23 in various histological subgroups of TGCTs and normal testes. The expression levels of miR-142-3p and PTPN23 in TGCTs (S1-S6) and normal testis samples (N1, N2) were validated by qRT-PCR. The expression of miR-142-3p in TGCTs was significantly higher than that in normal testicular sections (Fig. 6A). Remarkably, three of six cases displayed 5 times more expression of miR-142-3p. Thus, at least 50% of TGCTs express a very high level of miR-142-3p. In contrast, PTPN23 expression was significantly lower in TGCTs than in normal testes (Fig. 6A). In situ hybridization and immunohistochemical analyses confirmed that miR-142-3p expression levels were inversely correlated with PTPN23 protein levels in both TGCTs and normal testes (Fig. 6, B and C). We did not observe any differences between histological subgroups of TGCTs.
FIGURE 6.
PTPN23 is down-regulated significantly, and its expression levels correlates negatively with those of miR-142-3p in TGCTs and normal testes. A, expression of miR-142-3p (top panel) and PTPN23 (bottom panel) RNAs in human normal testes and TGCTs. RNU6B or GAPDH was used to normalize the amount of template cDNA. Experiments were repeated three times, and each value represents the mean ± S.E. B, expression of miR-142-3p RNA and PTPN23 protein in human normal testis (N1) and TGCTs. S1 and S3, seminoma; EC, embryonal carcinoma; YS, yolk sac tumor. Paraffin-embedded sections of human TGCTs and normal testicular regions were subjected to in situ hybridization with a digoxigenin-labeled miR-142-3p miRCURY LNA detection probe or to immunohistochemical analysis using an anti-PTPN23 antibody. Scale bar = 50 μm. Inset of left S1 panel, high-magnification view of the positive area. C, expression of PTPN23 in human normal testes. Sections from human normal testicular tissues (TE803) were subjected to immunohistochemical analysis of PTPN23 expression. Scale bar = 100 μm.
DISCUSSION
The PTPN23 gene is localized at human chromosome 3p21, a region known to be a hot spot for deletion in various carcinomas. This implicates the possibility for PTPN23 to function as a tumor suppressor. In this report, we showed that PTPN23 expression was down-regulated in human TGCT cell lines and primary tissues isolated from TGTC patients when compared with spermatogonial stem cells and normal testes samples.
We investigated the roles of PTPN23 in TGCT development. Because the serum requirement was increased by PTPN23 overexpression in NEC8 cells, we performed anchorage-independent growth assays and in vivo tumorigenesis assays. The results revealed that PTPN23 expression results in the suppression of neoplastic phenotypes. It has been reported that suppression of PTPN23 enhances the motility of endothelial cells and bladder carcinoma cells by modulating focal adhesion kinase (7, 26). Furthermore, suppression of PTPN23 induced mammary epithelial cell invasion and increased SRC activity (11). Cell migration capacity is critical for invasion and metastasis of cancer cells. In this report, we showed for the first time that PTPN23 strongly suppresses the tumorigenic activity of TGCTs. Although most critical tumor-suppressive functions of PTPN23 remain elusive, a higher serum requirement could contribute to the loss of tumorigenic activity in PTPN23/NEC8 cells.
Because tumor suppressor genes are recessive, both copies of a tumor suppressor gene should be inactivated by mutation or deletion to promote the tumor development. A mutational analysis of PTPs identified missense, nonsense, and frameshift mutations in colorectal carcinomas (27). They examined the coding exons of 87 members of the PTP family, including PTPN23, and detected 77 mutations in the PTPRF, PTPRG, PTPRT, PTPN3, PTPN13, and PTPN14 genes. Korff et al. (28) examined frameshift mutations in the coding repeats of 16 PTPs in 54 primary colorectal carcinomas and found microsatellite instability. Frameshift mutations in PTPN21 and PTPRS occurred with high frequency (16 and 12%, respectively). However, only one mutation was detected in an upstream region of the protein-tyrosine phosphatase domain of PTPN23 in colorectal carcinoma. Mutational analysis of PTPN23 in the Catalogue of Somatic Mutations in Cancer (COSMIC), Sanger Institute, identified 30 mutations in 5277 carcinomas. Mutations were identified in lung (10 samples), large intestine (9 samples), skin (3 samples), breast (3 samples), prostate (2 samples), central nervous system (1 sample), esophagus (1 sample), and upper aerodigestive tract (1 sample). However, no commonly mutated hot spot codons were detected. In two TGCT cell lines (NEC8 and NEC14) used in this study, we did not detect any mutations in the coding regions of the PTPN23 gene. Therefore, the loss of PTPN23 expression in TGCTs may be caused by different mechanisms.
It has been reported recently that oncogenic miRNAs suppress tumor suppressor genes and promote T cell acute lymphoblastic leukemia (29). We used multiple computational algorithms to search for miRNAs predicted to suppress PTPN23 expression and identified eight conserved nucleotides that matched the seed region of miR-142-3p within the 3′ UTR of the PTPN23 gene. Reporter assays proved that PTPN23 is a target of miR-142-3p. Moreover, ectopic expression of miR-142 in PTPN23/NEC8 cells efficiently repressed PTPN23 expression, thereby restoring the colony forming capacity in soft agar. Thus, a higher level of miR-142-3p is supportive for TGCTs by way of PTPN23 down-regulation. This is a novel molecular interaction between miRNA and PTPs during tumorigenesis.
We next found a significantly higher expression of miR-142-3p in TGCTs compared with normal testes. Differences in miR-142-3p expression between seminoma and non-seminoma samples were not detectable by in situ hybridization. In contrast, expression of PTPN23 in normal testicular tissues was significantly higher than that in TGCTs. miR-142-3p is an evolutionarily conserved miRNA that is expressed aberrantly in adult T cell leukemia (30), head and neck squamous cell carcinomas (31, 32), esophageal squamous cell carcinoma (33), lung adenocarcinoma (34), and hepatocellular carcinoma (35). These data suggest that miR-142-3p is a potential marker for the malignant progression of tumors and functions as an oncogenic miRNA in human cancers. In relation to this observation, a recent report indicated that miR-372 and miR-373 are highly expressed in TGCTs and inhibit the expression of the LATS2 tumor suppressor gene (24). Further, comparison of miRNA expression in normal testis and seminomas identified miR-373 as the most down-regulated miRNA among the 156 miRNAs present in normal testis. Interestingly, miR-142-3p was listed as the second most down-regulated miRNA (35). RAC1 and TAB2 were identified as target genes for miR-142-3p in hepatocellular carcinoma cells and acute myeloid leukemia (36, 37). RAC1, TAB2, and PTPN23 are in the high rank of 331 predicted targets (Target Scan Human Database, Release 6.2) and 102 conserved targets (PicTar Database) of miR-142-3p. These results strongly support PTPN23 as one of the target genes of miR-142-3p. miR-142-3p was expressed at high levels in TGCTs but at low to undetectable levels in normal testes, indicating that miR-142-3p might possess an oncogenic function in germ cell development. Many miRNAs are significantly overexpressed in various cancers. These oncogenic miRNAs functions in tumor development via repressing the expression of genes having tumor suppressor activity (38, 39).
However, the regulatory mechanisms that control the specific expression of miRNAs in human cancers are largely unknown. miR-142 is highly expressed in hematopoietic cells and directly targets the 3′UTR of the IL-6 gene (40).
Recently it was reported that PU.1, C/EBPβ, CBPβ, and Runx1 regulate the hematopoietic-specific expression of miR-142 (41). The transcriptional start sites of human miRNAs were reportedly identified on the basis of high-throughput sequencing data (42). A putative transcription start site for hsa-miR-142 is located 1200 bp upstream of the miR-142 precursor gene, and a putative TATA box is located 30 bp upstream of the transcription start site. We reported previously that a member of DEAD box protein family, Ddx1, is required for the testicular tumorigenesis and acts partially through the transcriptional activation of 12p stem cell genes (25). Interestingly, candidate binding sites for Ddx1-mediated transcriptional activation are present 290–310, 505–525, and 885–905 bp upstream of the transcription start site of the miR-142 precursor gene. Therefore, Ddx1 might be involved in the up-regulation of miR-142-3p in TGCTs. A future challenge will be to elucidate the miRNA-mediated regulatory networks in TGCTs.
In conclusion, we demonstrated that PTPN23 has a tumor suppressor activity in TGCTs. We also showed that miR-142-3p inhibits PTPN23 expression via a conserved miR-142-3p recognition motif located within the 3′ UTR of PTPN23 mRNA. Importantly, the expression of miR-142-3p correlated inversely with the PTPN23 level in TGCTs and normal testes. These data collectively reveal a novel miRNA-dependent down-regulation of PTPN23 expression that is important for the development of TGCTs.
Acknowledgments
We thank all stem cell project members for discussions and Dr. Noriko Nara (Yokohama City University Medical Center) for advice.
This study was supported by Japan Society for the Promotion of Science Grants-in-Aid (C) 23592356 (to K. T.) and (B) 23390256 (to T. H.) and Exploratory Research Grant 23659481 (to T. H.).
- PTP
- protein-tyrosine phosphatase
- TGCT
- testicular germ cell tumor
- miRNA
- microRNA
- qRT-PCR
- quantitative RT-PCR.
REFERENCES
- 1. Tanaka K., Tamura H., Tanaka H., Katoh M., Futamata Y., Seki N., Nishimune Y., Hara T. (2002) Spermatogonia-dependent expression of testicular genes in mice. Dev. Biol. 246, 466–479 [DOI] [PubMed] [Google Scholar]
- 2. Hendriks W. J., Elson A., Harroch S., Stoker A. W. (2008) Protein tyrosine phosphatases. Functional inferences from mouse models and human diseases. FEBS J. 275, 816–830 [DOI] [PubMed] [Google Scholar]
- 3. Scrima M., De Marco C., De Vita F., Fabiani F., Franco R., Pirozzi G., Rocco G., Malanga D., Viglietto G. (2012) The nonreceptor-type tyrosine phosphatase PTPN13 is a tumor suppressor gene in non-small cell lung cancer. Am. J. Pathol. 180, 1202–1214 [DOI] [PubMed] [Google Scholar]
- 4. Toyooka S., Ouchida M., Jitsumori Y., Tsukuda K., Sakai A., Nakamura A., Shimizu N., Shimizu K. (2000) HD-PTP. A novel protein tyrosine phosphatase gene on human chromosome 3p21.3. Biochem. Biophys. Res. Commun. 278, 671–678 [DOI] [PubMed] [Google Scholar]
- 5. Gingras M. C., Kharitidi D., Chénard V., Uetani N., Bouchard M., Tremblay M. L., Pause A. (2009) Expression analysis and essential role of the putative tyrosine phosphatase His-domain-containing protein tyrosine phosphatase (HD-PTP). Int. J. Dev. Biol. 53, 1069–1074 [DOI] [PubMed] [Google Scholar]
- 6. Cao L., Zhang L., Ruiz-Lozano P., Yang Q., Chien K. R., Graham R. M., Zhou M. (1998) A novel putative protein-tyrosine phosphatase contains a BRO1-like domain and suppresses Ha-ras-mediated transformation. J. Biol. Chem. 273, 21077–21083 [DOI] [PubMed] [Google Scholar]
- 7. Castiglioni S., Maier J. A., Mariotti M. (2007) The tyrosine phosphatase HD-PTP. A novel player in endothelial migration. Biochem. Biophys. Res. Commun. 364, 534–539 [DOI] [PubMed] [Google Scholar]
- 8. Mariotti M., Castiglioni S., Garcia-Manteiga J. M., Beguinot L., Maier J. A. (2009) HD-PTP inhibits endothelial migration through its interaction with Src. Int. J. Biochem. Cell Biol. 41, 687–693 [DOI] [PubMed] [Google Scholar]
- 9. Gingras M. C., Zhang Y. L., Kharitidi D., Barr A. J., Knapp S., Tremblay M. L., Pause A. (2009) HD-PTP is a catalytically inactive tyrosine phosphatase due to a conserved divergence in its phosphatase domain. PLoS ONE 4, e5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kim J., Lee J. E., Heynen-Genel S., Suyama E., Ono K., Lee K., Ideker T., Aza-Blanc P., Gleeson J. G. (2010) Functional genomic screen for modulators of ciliogenesis and cilium length. Nature 464, 1048–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lin G., Aranda V., Muthuswamy S. K., Tonks N. K. (2011) Identification of PTPN23 as a novel regulator of cell invasion in mammary epithelial cells from a loss-of-function screen of the “PTP-ome”. Genes Dev. 25, 1412–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Castiglioni S., Maier J. A. (2012) The tyrosine phosphatase HD-PTP (PTPN23) is degraded by calpains in a calcium-dependent manner. Biochem. Biophys. Res. Commun. 421, 380–383 [DOI] [PubMed] [Google Scholar]
- 13. Foster K., Osborne R. J., Huddart R. A., Affara N. A., Ferguson-Smith M. A., Maher E. R. (1995) Molecular genetic analysis of the von Hippel-Lindau disease (VHL) tumour suppressor gene in gonadal tumours. Eur. J. Cancer 31A, 2392–2395 [DOI] [PubMed] [Google Scholar]
- 14. Lothe R. A., Fosså S. D., Stenwig A. E., Nakamura Y., White R., Børresen A. L., Brøgger A. (1989) Loss of 3p or 11p alleles is associated with testicular cancer tumors. Genomics 5, 134–138 [DOI] [PubMed] [Google Scholar]
- 15. Eyzaguirre E., Gatalica Z. (2002) Loss of Fhit expression in testicular germ cell tumors and intratubular germ cell neoplasia. Mod. Pathol. 15, 1068–1072 [DOI] [PubMed] [Google Scholar]
- 16. Sekido Y., Ahmadian M., Wistuba I. I., Latif F., Bader S., Wei M. H., Duh F. M., Gazdar A. F., Lerman M. I., Minna J. D. (1998) Cloning of a breast cancer homozygous deletion junction narrows the region of search for a 3p21.3 tumor suppressor gene. Oncogene 16, 3151–3157 [DOI] [PubMed] [Google Scholar]
- 17. Wei M. H., Latif F., Bader S., Kashuba V., Chen J. Y., Duh F. M., Sekido Y., Lee C. C., Geil L., Kuzmin I., Zabarovsky E., Klein G., Zbar B., Minna J. D., Lerman M. I. (1996) Construction of a 600-kilobase cosmid clone contig and generation of a transcriptional map surrounding the lung cancer tumor suppressor gene (TSG) locus on human chromosome 3p21.3. Progress toward the isolation of a lung cancer TSG. Cancer Res. 56, 1487–1492 [PubMed] [Google Scholar]
- 18. Cheng Y., Poulos N. E., Lung M. L., Hampton G., Ou B., Lerman M. I., Stanbridge E. J. (1998) Functional evidence for a nasopharyngeal carcinoma tumor suppressor gene that maps at chromosome 3p21.3. Proc. Natl. Acad. Sci. U.S.A. 95, 3042–3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wistuba I. I., Montellano F. D., Milchgrub S., Virmani A. K., Behrens C., Chen H., Ahmadian M., Nowak J. A., Muller C., Minna J. D., Gazdar A. F. (1997) Deletions of chromosome 3p are frequent and early events in the pathogenesis of uterine cervical carcinoma. Cancer Res. 57, 3154–3158 [PubMed] [Google Scholar]
- 20. Dreijerink K., Braga E., Kuzmin I., Geil L., Duh F. M., Angeloni D., Zbar B., Lerman M. I., Stanbridge E. J., Minna J. D., Protopopov A., Li J., Kashuba V., Klein G., Zabarovsky E. R. (2001) The candidate tumor suppressor gene, RASSF1A, from human chromosome 3p21.3 is involved in kidney tumorigenesis. Proc. Natl. Acad. Sci. U.S.A. 98, 7504–7509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Meng F., Henson R., Wehbe-Janek H., Ghoshal K., Jacob S. T., Patel T. (2007) MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology 133, 647–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ventura A., Young A. G., Winslow M. M., Lintault L., Meissner A., Erkeland S. J., Newman J., Bronson R. T., Crowley D., Stone J. R., Jaenisch R., Sharp P. A., Jacks T. (2008) Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell 132, 875–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Garofalo M., Di Leva G., Romano G., Nuovo G., Suh S. S., Ngankeu A., Taccioli C., Pichiorri F., Alder H., Secchiero P., Gasparini P., Gonelli A., Costinean S., Acunzo M., Condorelli G., Croce C. M. (2009) miR-221 and 222 regulate TRAIL resistance and enhance tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell 16, 498–509 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24. Voorhoeve P. M., le Sage C., Schrier M., Gillis A. J., Stoop H., Nagel R., Liu Y. P., van Duijse J., Drost J., Griekspoor A., Zlotorynski E., Yabuta N., De Vita G., Nojima H., Looijenga L. H., Agami R. (2006) A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell 124, 1169–1181 [DOI] [PubMed] [Google Scholar]
- 25. Tanaka K., Okamoto S., Ishikawa Y., Tamura H., Hara T. (2009) DDX1 is required for testicular tumorigenesis, partially through the transcriptional activation of 12p stem cell genes. Oncogene 28, 2142–2151 [DOI] [PubMed] [Google Scholar]
- 26. Mariotti M., Castiglioni S., Maier J. A. (2009) Inhibition of T24 human bladder carcinoma cell migration by RNA interference suppressing the expression of HD-PTP. Cancer Lett. 273, 155–163 [DOI] [PubMed] [Google Scholar]
- 27. Wang Z., Shen D., Parsons D. W., Bardelli A., Sager J., Szabo S., Ptak J., Silliman N., Peters B. A., van der Heijden M. S., Parmigiani G., Yan H., Wang T. L., Riggins G., Powell S. M., Willson J. K., Markowitz S., Kinzler K. W., Vogelstein B., Velculescu V. E. (2004) Mutational analysis of the tyrosine phosphatome in colorectal cancers. Science 304, 1164–1166 [DOI] [PubMed] [Google Scholar]
- 28. Korff S., Woerner S. M., Yuan Y. P., Bork P., von Knebel Doeberitz M., Gebert J. (2008) Frameshift mutations in coding repeats of protein tyrosine phosphatase genes in colorectal tumors with microsatellite instability. BMC Cancer 8, 329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mavrakis K. J., Van Der Meulen J., Wolfe A. L., Liu X., Mets E., Taghon T., Khan A. A., Setty M., Setti M., Rondou P., Vandenberghe P., Delabesse E., Benoit Y., Socci N. B., Leslie C. S., Van Vlierberghe P., Speleman F., Wendel H. G. (2011) A cooperative microRNA-tumor suppressor gene network in acute T-cell lymphoblastic leukemia (T-ALL). Nat. Genet. 43, 673–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bellon M., Lepelletier Y., Hermine O., Nicot C. (2009) Deregulation of microRNA involved in hematopoiesis and the immune response in HTLV-I adult T-cell leukemia. Blood 113, 4914–4917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chang S. S., Jiang W. W., Smith I., Poeta L. M., Begum S., Glazer C., Shan S., Westra W., Sidransky D., Califano J. A. (2008) MicroRNA alterations in head and neck squamous cell carcinoma. Int. J. Cancer 123, 2791–2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hui A. B., Lenarduzzi M., Krushel T., Waldron L., Pintilie M., Shi W., Perez-Ordonez B., Jurisica I., O'Sullivan B., Waldron J., Gullane P., Cummings B., Liu F. F. (2010) Comprehensive microRNA profiling for head and neck squamous cell carcinomas. Clin. Cancer Res. 16, 1129–1139 [DOI] [PubMed] [Google Scholar]
- 33. Lin R. J., Xiao D. W., Liao L. D., Chen T., Xie Z. F., Huang W. Z., Wang W. S., Jiang T. F., Wu B. L., Li E. M., Xu L. Y. (2012) MiR-142-3p as a potential prognostic biomarker for esophageal squamous cell carcinoma. J. Surg. Oncol. 105, 175–182 [DOI] [PubMed] [Google Scholar]
- 34. Kaduthanam S., Gade S., Meister M., Brase J. C., Johannes M., Dienemann H., Warth A., Schnabel P. A., Herth F. J., Sültmann H., Muley T., Kuner R. (2013) Serum miR-142-3p is associated with early relapse in operable lung adenocarcinoma patients. Lung Cancer 80, 223–227 [DOI] [PubMed] [Google Scholar]
- 35. Gillis A. J., Stoop H. J., Hersmus R., Oosterhuis J. W., Sun Y., Chen C., Guenther S., Sherlock J., Veltman I., Baeten J., van der Spek P. J., de Alarcon P., Looijenga L. H. (2007) High-throughput microRNAome analysis in human germ cell tumours. J. Pathol. 213, 319–328 [DOI] [PubMed] [Google Scholar]
- 36. Wang X. S., Gong J. N., Yu J., Wang F., Zhang X. H., Yin X. L., Tan Z. Q., Luo Z. M., Yang G. H., Shen C., Zhang J. W. (2012) MicroRNA-29a and microRNA-142-3p are regulators of myeloid differentiation and acute myeloid leukemia. Blood 119, 4992–5004 [DOI] [PubMed] [Google Scholar]
- 37. Wu L., Cai C., Wang X., Liu M., Li X., Tang H. (2011) MicroRNA-142–3p, a new regulator of RAC1, suppresses the migration and invasion of hepatocellular carcinoma cells. FEBS Lett. 585, 1322–1330 [DOI] [PubMed] [Google Scholar]
- 38. Zhang B., Pan X., Cobb G. P., Anderson T. A. (2007) microRNAs as oncogenes and tumor suppressors. Dev. Biol. 302, 1–12 [DOI] [PubMed] [Google Scholar]
- 39. Esquela-Kerscher A., Slack F. J. (2006) Oncomirs. MicroRNAs with a role in cancer. Nat. Rev. Cancer 6, 259–269 [DOI] [PubMed] [Google Scholar]
- 40. Sun Y., Varambally S., Maher C. A., Cao Q., Chockley P., Toubai T., Malter C., Nieves E., Tawara I., Wang Y., Ward P. A., Chinnaiyan A., Reddy P. (2011) Targeting of microRNA-142-3p in dendritic cells regulates endotoxin-induced mortality. Blood 117, 6172–6183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sun Y., Sun J., Tomomi T., Nieves E., Mathewson N., Tamaki H., Evers R., Reddy P. (2013) PU1-dependent transcriptional regulation of miR-142 contributes to its hematopoietic cell-specific expression and modulation of IL-6. J. Immunol. 190, 4005–4013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chien C. H., Sun Y. M., Chang W. C., Chiang-Hsieh P. Y., Lee T. Y., Tsai W. C., Horng J. T., Tsou A. P., Huang H. D. (2011) Identifying transcriptional start sites of human microRNAs based on high-throughput sequencing data. Nucleic Acids Res. 39, 9345–9356 [DOI] [PMC free article] [PubMed] [Google Scholar]