FIGURE 1.
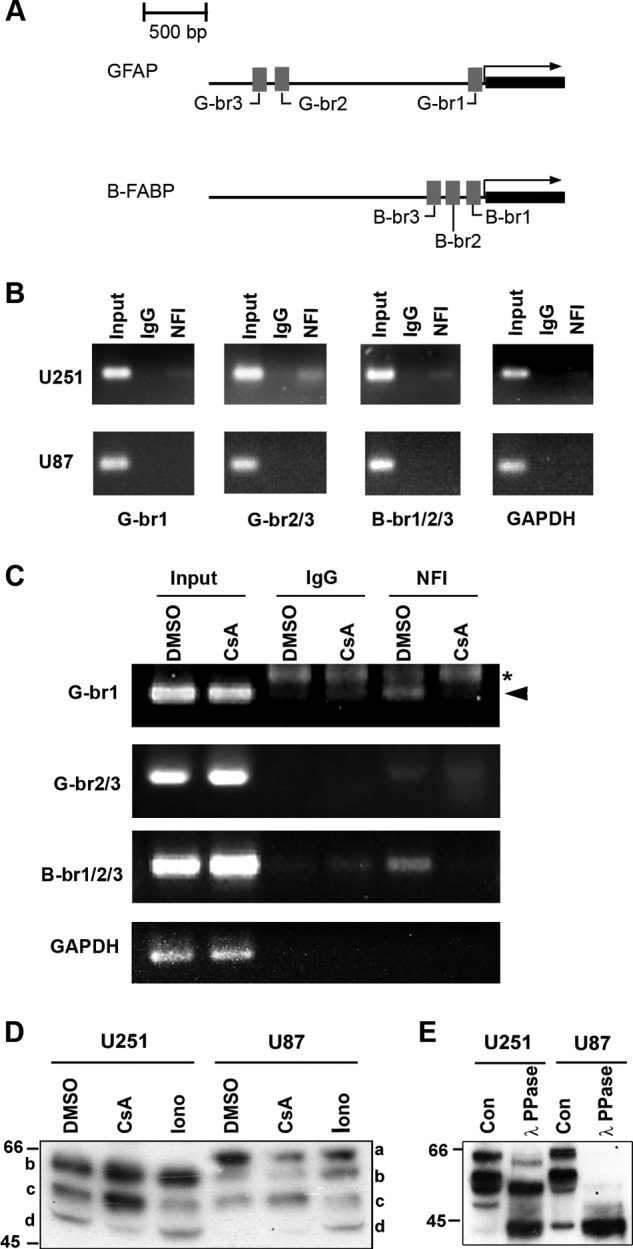
NFI-dependent promoter binding and activity. A, schematic diagram of the GFAP and B-FABP promoter regions showing the relative location of the three NFI-binding sites located in each promoter. B, ChIP analysis was carried out in U251 and U87 cells using either a pan-specific anti-NFI antibody or normal rabbit IgG as a negative control. Primers flanking the NFI-binding sites identified in the GFAP (G-br1 and G-br2/3) and B-FABP promoters (B-br1/2/3) were used for PCR amplification (Table 1). Primers flanking the proximal GAPDH promoter were used as a negative control. Input DNA represents DNA isolated from U251 or U87 cell lysates following sonication but prior to immunoprecipitation. Input DNA serves as the positive control for the PCR and reveals products of the expected sizes. C, U251 cells were treated for 1 h with 1 μm CsA or DMSO (control), followed by ChIP analysis as described in B. Asterisk denotes a nonspecific band, and arrowhead denotes a specific band. D, Western blot analysis of NFI in U251 and U87 cells treated with CsA and ionomycin (Iono). U251 and U87 cells were treated for 1 h with DMSO, 1 μm CsA, or 1 μm ionomycin and harvested using trypsin to dissociate the cells. Nuclear extracts were electrophoresed in a 10% SDS-polyacrylamide gel, transferred to a PVDF membrane, and immunostained with rabbit anti-NFI antibody. The primary antibody was detected with horseradish peroxidase-conjugated antibody, and the signal was detected with Immobilon reagent. E, λ-phosphatase treatment of U251 and U87 nuclear extracts. U251 and U87 nuclear extracts were prepared in the absence of phosphatase inhibitors and treated with or without λ-phosphatase (PPase) for 1 h at 30 °C. Following treatment, extracts were subjected to Western blot analysis as in D. Con, control.
