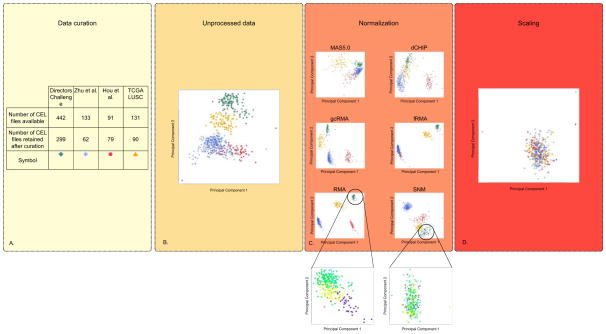
Figure 1.
Overview of the pre-processing framework. Effects on the structure of the data are represented by principle component plots for four NSCLC gene expression datasets processed separately. (A) A Table to represent the number of raw data (CEL files) included in study as a result of the data curation process. As the classifier is for early-stage patients, an explicit decision was made to only include those who are pathological stage IA to IIIA, who did not receive induction or adjuvant chemotherapy and patients for whom overall survival (OS) data are available. In addition, only patients who underwent complete tumor resection were included. Finally, gene expression outliers were identified graphically and removed from further analyses. (B) Unprocessed data analyzed by principal component analysis plot (C) Effect of five widely used unsupervised (RMA (Robust Multi-array Average) (71), gcRMA (GC Robust Multi-array Average) (72), MAS5.0 (Affymetrix Multiarray Suite 5.0) (73), dCHIP (DNA Chip Analyzer) (74) and fRMA (frozen Robust Multiarray Analysis) (75)) and one supervised (SNM) (14) normalization methods on the structure of the data. The effect of normalization on only the patients included in the Directors Challenge dataset are shown in the callouts from SNM and RMA with batch represented by different colors on principal component plot. (D) Principal component plot of SNM normalized data normalized to unit variance and 0 mean. The data and code to generate these plots are made available (see supplementary material).