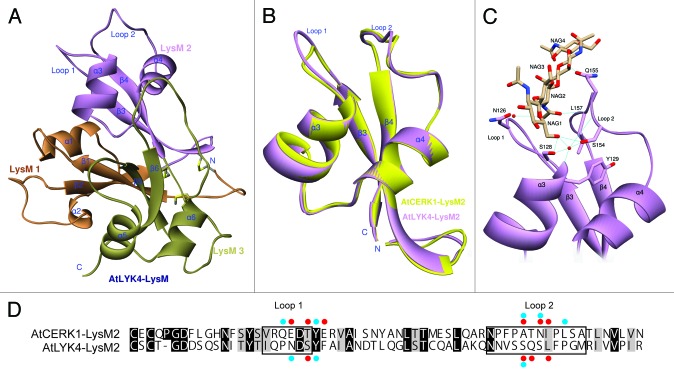
Figure 2. Three-dimensional model prediction of the structure of the AtLYK4 extracellular domain, including a ligand docking model. (A)The 3D model of AtLYK4 extracellular domain was built based on the crystal structure of AtCERK1 (PDB code: 4EBY). Each LysM domain is represented in a different color: first LysM (orange), second LysM (purple) and third LysM (green). (B)The second LysM domains of AtLYK4 (purple) and AtCERK1 (yellow) are superimposed to highlight the similarity in structure. Note that the overall folds are highly conserved between the two models, although the AtLYK4 has the longer extended Loop 1 which is a constitutive part of the cleft where the predicted chitin-binding site is found. (C) Docking model between the second LysM domain of AtLYK4 and chitotetraose. (D) Pairwise sequence comparison of the second LysM domains of AtLYK4 and AtCERK1. Identical and similar residues throughout the alignment are shown in black and gray, respectively. Enclosed boxes represent Loop 1 and Loop 2. Red and blue dots indicate the residues involved in direct interactions and water molecule-mediated interactions with chitotetraose, respectively. The residues involved in van der Waals interactions were neglected here.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.