Abstract
Green foxtail (Setaria viridis) is a new model plant for the genomic investigation of C4 photosynthesis biology. As the ancestor of foxtail millet (Setaria italica), an ancient cereal of great importance in arid regions of the world, green foxtail is crucial for the study of domestication and evolution of this ancient crop. In the present study, 288 green foxtail accessions, which were collected from all geographical regions of China, were analysed using 77 simple sequence repeats (SSRs) that cover the whole genome. A high degree of molecular diversity was detected in these accessions, with an average of 33.5 alleles per locus. Two clusters, which were inconsistent with the distribution of eco-geographical regions in China, were inferred from STRUCTURE, Neighbor–Joining, and principal component analysis, indicating a partially mixed distribution of Chinese green foxtails. The higher subpopulation diversity was from accessions mainly collected from North China. A low level of linkage disequilibrium was observed in the green foxtail genome. Furthermore, a combined analysis of green foxtail and foxtail millet landraces was conducted, and the origin and domestication of foxtail millet was inferred in North China.
Key words: Genetic diversity, germplasm, green foxtail, Setaria viridis.
Introduction
Green foxtail, Setaria viridis (L.) Beauv., also called green millet, belongs to Setaria Beauv., a member of the grass tribe Paniceae in the subfamily Panicoideae. Green foxtail is the ancestor of cultivated foxtail millet (Setaria italica L. Beauv.), an ancient cereal of great importance to dry land agriculture that has been grown in China for >10,500 years (Yang et al., 2012) for grain and forage. Due to its small growth stature, small genome size, self-fertilization, short growing cycle, and efficient genetic transformation, green foxtail is being rapidly developed as a model plant for deciphering C4 photosynthesis biology which was verified to be highly efficient in CO2 fixation and has great potential for the genetic improvement of C3 staple food crops such as rice (Brutnell et al., 2010; Li and Brutnell, 2011; Bennetzen et al., 2012). Green foxtail is closely related to biofuel grasses such as switch grass (Panicum virgatum L.), proso millet (Panicum miliaceum L.), and pearl millet (Pennisetum glaucum L.). The genomic and genetic annotation of green foxtail will undoubtedly improve breeding programmes in those crops which are difficult to manipulate due to their large genomes, outcrossing breeding systems, large stature, and long growing cycles (Doust et al., 2009; Bennetzen et al., 2012). Foxtail millet is also rapidly becoming a model plant for the functional genomics of grass, focusing on crop domestication, abiotic stress tolerance, and grass evolution (Doust et al., 2009; Li and Brutnell, 2011; Lata et al., 2012; Mauro-Herrera et al., 2013). The release of the draft genome sequences of foxtail millet has accelerated the development of green foxtail and foxtail millet as a novel model system (Bennetzen et al., 2012; Zhang et al., 2012).
Green foxtail is an Old World species of the Setaria genus and it is now distributed worldwide (Austin, 2006). As a regional crop, wild germplasm collections of foxtail millet and related studies have been sparse (Li and Brutnell, 2011). Wang et al. (1995) investigated the genetic diversity and structure of green foxtail accessions collected from North America and Eurasia using allozyme markers, and suggested that there were similar isozymatic forms between foxtail millet and green foxtail collected from the same regions. Chinese accessions were identified as having a high level of genetic diversity using amplified fragment length polymorphism (AFLP) markers (Le Thierry d’Ennequin et al., 2000), transposon display (Hirano et al., 2011), and intersimple sequence repeat (ISSR) markers (Li et al., 2012), and the well known A10 accession which was used as a model for C4 photosynthesis study was also from China (Brutnell et al., 2010; Li and Brutnell, 2011; Bennetzen et al., 2012; Caemmerer et al., 2012). However, all these studies were carried out with a small sample size of no more than 40 accessions. Detailed information regarding the genetic diversity and population structure of green foxtail with a larger sample size will be useful in many related studies.
The genetic diversity and population structure of both the wild relatives and domesticated landraces have been widely used for crop origin studies (Spooner et al., 2005; Heerwaarden et al., 2011; Huang et al., 2012). From studies on wild green foxtail and cultivated foxtail millet, monotopic (Fukunaga et al., 2006; Hirano et al., 2011; Li et al., 2012) and polytopic (Jusuf and Pernes, 1985) origins of foxtail millet have been inferred, and most reports agree that China was the first site of foxtail millet domestication, if not the only one (Vavilov, 1926; Austin, 2006; Li et al., 2012). This is supported not only by the earliest archeological evidence of foxtail millet identified in Northern China, which existed >10 500 years ago (Lu et al., 2009; Barton et al., 2009; Yang et al., 2012), but also by the high level of genetic diversity of Chinese foxtail millet landraces (Le Thierry d’Ennequin et al., 2000; Fukunaga et al., 2006; Hirano et al., 2011). The analysis of genetic diversity and population structure based on a large sample size of 250 Chinese foxtail millet landraces, which represent 1% of the foxtail millet germplasm kept in the Chinese National Gene Bank, revealed a high genetic diversity of 20.9 alleles per locus, and classified the accessions into four subpopulations, in accordance with its eco-geographical distribution in China (Wang et al., 2012). The diversity analysis also suggested that foxtail millet was first domesticated in the Yellow River drainage area (E95°53′–119°05′, N32°10′–41°50′, including Gansu, Shannxi, Shanxi, Henan, and Hebei) and then spread to other parts of the country. Investigation of the genetic diversity and population genetics of Chinese green foxtail can be used to provide complementary information, enabling a clearer understanding of this issue.
Association mapping is an effective approach requiring information on population structure and linkage disequilibrium (LD) to detect quantitative trait loci (QTLs)/genes of great importance. Using simple sequence repeat (SSR) markers, association mapping has been successfully developed in rice (Jin et al., 2010), wheat (Kruger et al., 2004; Maccaferri et al., 2005), and barley (Mather et al., 2004). However, molecular genetic investigations of the diversity, population structure, and LD patterns of green foxtail using SSR markers have not yet been performed.
In this study, a large sample size of 288 green foxtail accessions collected from all geographical regions of China were analysed using SSR markers covering the nine chromosomes of green foxtail. Genetic diversity and population structure were inferred by software simulations. LD levels in the green foxtail genome were also measured. An analysis of data on green foxtail combined with previous data on foxtail millet landraces (Wang et al., 2012) was also carried out and the results were compared. The data and conclusions of this study will benefit green foxtail germplasm collection and management, genomic studies, trait association mapping, and breeding applications.
Materials and methods
Green foxtail sampling
All of the green foxtail samples were collected from China in 2010. The green foxtail samples used in this study were selected from different eco-regions, and the number of accessions sampled from each region was in proportion to the number of foxtail millet accessions stored in the Chinese National Gene Bank (CNGB) from each region. Thus, the number of green foxtail samples from Northern China, where foxtail millet has a large growing area and there are many collected accessions, was larger than that of Southern China. The aim of the sampling strategy was to assemble a more representative set of accessions of green foxtail from all the eco-regions of China. The number of samples from each province used in this study is listed in Table 1.
Table 1.
Origin of green foxtail selected in this trial
| Eco-regions of foxtail millet | Province | No. of accessions |
|---|---|---|
| Early spring-sowing region (ESR) | Heilongjiang | 12 |
| Spring-sowing region (SR) | Shanxi | 39 |
| Shannxi | 22 | |
| Gansu | 18 | |
| Inner Mongolia | 5 | |
| North Hebei | 24 | |
| Tibet | 2 | |
| Xinjiang | 5 | |
| Ningxia | 6 | |
| Summer- and spring-sowing region (SSSR) | Beijing | 6 |
| South Hebei | 13 | |
| Henan | 35 | |
| Shandong | 9 | |
| Tianjin | 1 | |
| Jilin | 12 | |
| Liaoning | 11 | |
| Southern China region (SCR) | Sichuan | 6 |
| Hubei | 9 | |
| Guangxi | 4 | |
| Guangdong | 9 | |
| Jiangsu | 9 | |
| Zhejiang | 5 | |
| Jiangxi | 10 | |
| Guizhou | 2 | |
| Fujian | 6 | |
| Yunnan | 8 |
Genotyping of green foxtail
The template DNA was extracted from leaves of the sampled green foxtail accessions using the cetyltrimethylammonium bromide (CTAB) method (Doyle, 1991). The 77 SSR markers described previously (Wang et al., 2012) were used. All SSRs were labelled with different coloured fluorescent dyes at the 5′ end of the forward primer for PCR amplification (Applied Biosystems, USA). The PCR mixture consisted of 1× Taq reaction buffer (Takala, with Mg2+), nucleotides dATP, dGTP, dCTP, and dTTP (125 μM each), 0.1 μM primer, 1U of Taq DNA polymerase, and 10ng of template DNA. The length of the amplified fragment of DNA was measured using an ABI 3730XL analyser. Polymorphism data were analysed using GeneMapper (Version 4.0). Microchecker 2.2.3 (Oosterhout et al., 2004) was used for checking mistakes due to potential primer stuttering to make sure the genotyping data were reliable.
Genetic diversity and population structure
All summary statistics such as allele number per locus, genotype number per locus, gene diversity, PIC (polymorphism information content) values, observed homozygosity, genetic distance, and F ST tests were determined using PowerMarker version 3.25 (Liu and Muse, 2005). Nei’s genetic distance (Nei and Takezaki, 1983) was calculated and used for unrooted phylogeny reconstruction based on Neighbor–Joining methods as implemented by PowerMarker software, and the tree was visualized using MEGA 5.0 (Tamura et al., 2007). Principal component analysis (PCA) was carried out in GenALEX 6.4 (Peakall and Smouse, 2006). Analysis of molecular variance (AMOVA) was calculated by PowerMarker. Three levels of AMOVA were conducted for each inferred subpopulation, including the molecular variance of ‘among two SSR alleles within individuals’, ‘among individuals within populations’, and ‘among populations’. The lengths of the amplified product of SSR markers of each accession were used as the value of microsatellite alleles for variance analysis.
The model-based software program STRUCTURE v2.3 (Pritchard et al., 2000) was used to infer population structure by a Bayesian approach using the SSR marker data set. The optimal value of K (the number of clusters) was deduced by evaluating K=1–14. Admixture and non-admixture models were used separately and allele frequencies were assumed to be correlated or independent in these two models, respectively. Length of burn-in of the Markov Chain Monte Carlo (MCMC) iterations was set to 100 000 and data were collected over 100 000 MCMC iterations in each run. Twenty iterations per K were conducted. The optimal value of K was identified using both the ad hoc procedure introduced by Pritchard et al. (2000) and the method developed by Evanno et al. (2005). Genetic diversity, private allele number, and divergence estimates were calculated for the different clusters identified by the structure analysis. Substructures within each main cluster were detected by the same approach using STRUCTURE v2.3.
Linkage disequilibrium
LD was evaluated for each pair of SSR loci by TASSEL, both on all accessions individually and on the clusters as inferred by STRUCTURE. D′ and r 2 LD measures modified for loci were used (Hedrick et al., 1987; Weir et al., 1996). Significance (P-value) of D′ and r 2 for each SSR pair was determined by 100 000 permutations.
Bottleneck identification, gene flow estimation, candidate gene mining, and evolutionary analysis
Evidence for a bottleneck during the domestication of foxtail millet was obtained from data in this study and from previously published data (Wang et al., 2012), from parameters such as allele number, genotype number, gene diversity, and PIC value per locus. Gene flow within and between wild green foxtail and domesticated foxtail millet were inferred from F ST using Nm=0.25(1–F ST)/F ST (Slatkin et al., 1989). SSR loci under selection pressure between the two gene pools were identified by their higher (top 10) F ST values. SSR primer sequences were analysed using BLASTN tools in Phytozome v8.0 (http://www.phytozome.net/search.php), and annotated genes overlapping with SSR loci were selected as candidates. Neighbor–Joining trees of all accessions of wild green foxtail and domesticated foxtail millet were constructed for evolutionary analysis.
Results
Genetic diversity of green foxtail
Sixty-nine of the 77 markers were successfully amplified from the green foxtail accessions, and all the markers were polymorphic across the 288 green foxtail accessions. A total of 2312 alleles were detected, and the average allele number per locus was 33.50, ranging from 12 to 54. The average genotype number per locus was 46.37, ranging from 21 to 141. The average diversity of each locus was 0.90, ranging from 0.70 to 0.97. The PIC value for the markers was 0.90, ranging from 0.66 to 0.97. The average heterozygosity per locus was 0.07, ranging from 0.01 to 0.64. The average homozygosity extended to 0.90, which implies that the green foxtail samples are close to inbred lines (Table 2).
Table 2.
Genetic diversity of 288 green foxtail samples as revealed by SSR markers
| Sample | No. of alleles | No. of genotypes | Gene diversity | Heterozygosity | PIC |
|---|---|---|---|---|---|
| Average | 33.50 | 46.37 | 0.90 | 0.07 | 0.90 |
| Range | 12–54 | 21–141 | 0.70–0.97 | 0.01–0.64 | 0.66–0.97 |
| SD | 11.26 | 21.14 | 0.06 | 0.10 | 0.07 |
Population structure of green foxtail inferred by the admixture model
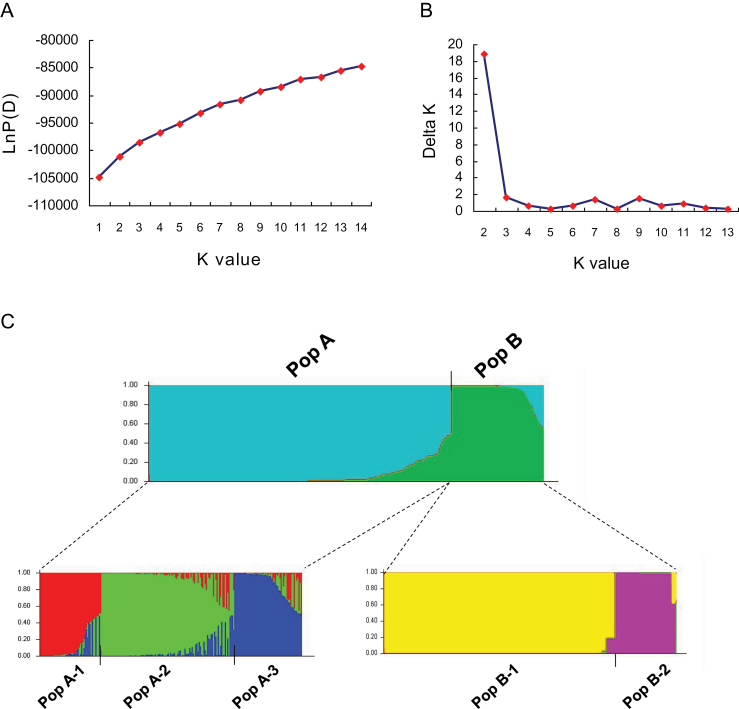
Admixture model-based calculations were conducted based on varying K from 1 to 14 with 20 iterations per K. When the STRUCTURE simulations were performed using all 288 accessions, the LnP(D) value increased with K from 1 to 14, but showed an evident inflection at K=2 (Fig. 1A). This result indicated that there might be two divergent subpopulations. According to the second-order statistics developed for STRUCTURE (Evanno et al., 2005) to estimate the number of subpopulations, the optimal value of K=2, at which the delta K showed a peak, was identified (Fig. 1B). This suggested that the Chinese green foxtail samples can be grouped into two populations, designated PopA and PopB. For each inferred population, substructuring under the topmost hierarchy was detected using a similar approach. PopA was divided into three (K=3) and PopB was divided into two (K=2) subgroups, with 20 iterations for each K (Supplementary Fig. S1A–D avalable at JXB online), making five subgroups in all. These were labelled as PopA-1 (63 samples), PopA-2 (107 samples), PopA-3 (51 samples), PopB-1 (51 samples), and PopB-2 (16 samples) (Fig. 1C, lower part).
Fig. 1.
Determination of the optimal value of K and inferred population structure of Chinese green foxtail accessions. (A) The ad hoc procedure described in Pritchard et al. (2000). (B) The second order of statistics (Delta K) based on Evanno et al. (2005). (C) Optimal population structure (K=2) and substructuring of PopA (K=3) and PopB (K=2). Each single vertical line represent a green foxtail sample, and different colours represents diverse clusters and subpopulations. The length of the coloured segment illustrates the green foxtail’s estimated proportion of membership in corresponding clusters as calculated through STRUCTURE.
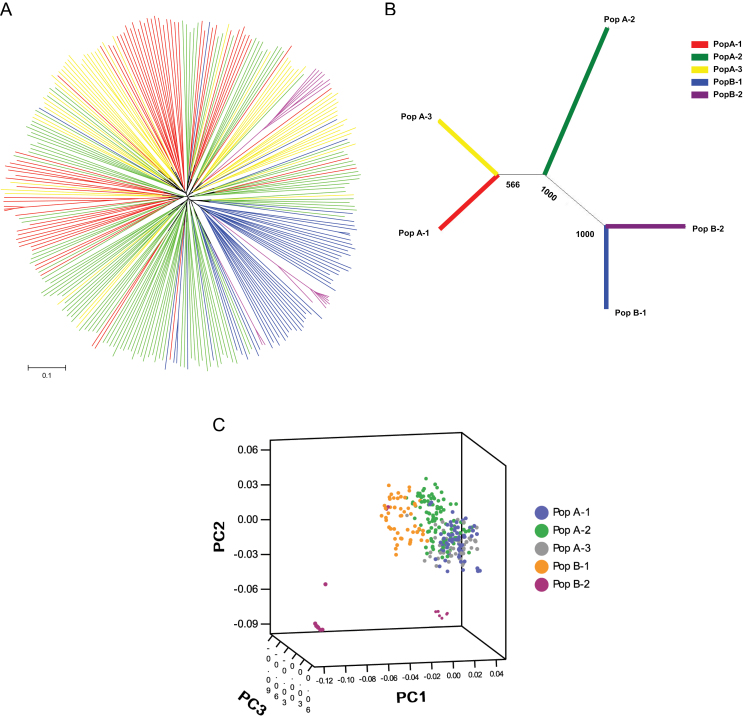
A Neighbor–Joining tree of 288 accessions was constructed based on Nei’s (Nei and Takezaki, 1983) genetic distance (Fig. 2A, B), which illustrated genetic relationships that closely approximated the STRUCTURE-based membership assignment for most of the accessions. The mixed distribution of the subgroup accessions into different clusters can be seen in Fig. 2A. A PCA was conducted to assess further the population subdivisions identified using STRUCTURE (Fig. 2C). The first principal component explained 24.66%, the second principal component explained 19.71%, and the third principal component explained 16.70% of the molecular marker variation among the 288 accessions. Plotting of the first three principal components showed separation of inferred subpopulations, which was highly consistent with STRUCTURE and the Neighbor–Joining analysis above.
Fig. 2.
Neighbor–Joining (NJ) analysis and principal component analysis (PCA) of Chinese green foxtail accessions. (A) Unrooted NJ tree of 288 green foxtail; every coloured branch represents one accession collected from the corresponding inferred subpopulation. (B) NJ tree of inferred subgroups based on Nei’s genetic distance; the bootstrap value (out of 1000) is indicated at the branch point. (C) Differentiation of genotypes from subpopulations according to the first principal component derived from diversity analysis of 69 SSR markers.
Relationships among subpopulations simulated from STRUCTURE were identified using pairwise genetic distance and F ST analysis (Table 3). The genetic distance ranged from 0.2420 between PopA-1 and PopA-2 to 0.6521 between PopA-3 and PopB-2, with an average of 0.4338. The pairwise F ST values for the subpopulations ranged from 0.0224 between PopA-1 and PopA-2 to 0.3474 between PopA-2 and B-2, with an average of 0.1301. The genetic distance was consistent with the trends of the F ST estimates. For instance, higher genetic distance and F ST were found between PopA and PopB clusters. The relationships of all five subpopulations suggested by Table 3 were also concordant with the Neighbor–Joining analysis including all 288 accessions.
Table 3.
Pairwise estimates of FST and genetic distance among five model-based subpopulations
| Subpopulation | PopA-1 | PopA-2 | PopA-3 | PopB-1 | PopB-2 |
|---|---|---|---|---|---|
| PopA-1 | 0.0224 | 0.0232 | 0.0568 | 0.2677 | |
| PopA-2 | 0.2420 | 0.0267 | 0.0532 | 0.3474 | |
| PopA-3 | 0.3196 | 0.2545 | 0.0572 | 0.2343 | |
| PopB-1 | 0.3792 | 0.2877 | 0.3926 | 0.2122 | |
| PopB-2 | 0.6518 | 0.5912 | 0.6521 | 0.5677 |
F ST estimates appear above the diagonal, and pairwise genetic distance appears below the diagonal.
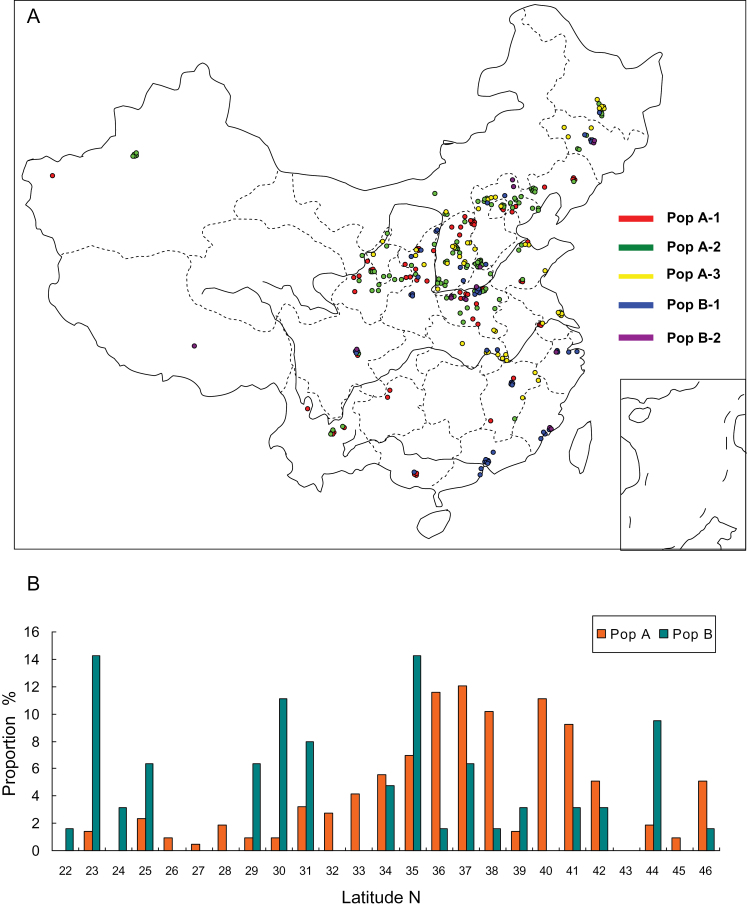
To interpret the separation among the subpopulations identified above, the habitat and geographical location where each accession was collected were depicted (Fig. 3A), but this did not reveal a clear pattern of ecological differentiation corresponding to the genetic subpopulations. Instead, spreading and mixing of the subpopulations throughout the pre-defined eco-regions were inferred. The proportion of individuals collected from each subgroup according to their latitude was also calculated (Fig. 3B). It was found that PopA included more accessions collected from Northern China and PopB contained a large proportion of individuals from Southern China.
Fig. 3.
Spatial distribution of collected green foxtail. (A) Map of the collection locations of Chinese green foxtail accessions grouped by five subpopulations inferred in this investigation. Different coloured spots represent individuals from the various subpopulations. (B) Proportions of individuals from corresponding subgroups based on latitudes of sampled locations.
Genetic variation of subpopulations
The genetic diversity per locus was estimated for each subpopulation (Tables 4 and 5). PopA-1 had the highest gene diversity and PIC value, followed by PopA-2 and PopA-3. The highest number of population-specific alleles was found in PopA-2. Among the 2312 alleles detected in the total populations, 627 (27.12%) were subpopulation-specific or private alleles. PopB-2 had the lowest genetic diversity identified in this research. PopA-2 had the largest genetic variance (34.62%) among all the subpopulations, followed by PopA-1 (20.69%), PopA-3 (16.42%), PopB-1 (15.57%), and PopB-2 (3.38%).
Table 5.
AMOVA of inferred subpopulations
| Source of variations | Sum of variances | Percentage | |
|---|---|---|---|
| Among individuals within populations | PopA-1 | 6672.9412 | 0.2069 |
| PopA-2 | 11165.3743 | 0.3462 | |
| PopA-3 | 5294.7049 | 0.1642 | |
| PopB-1 | 5022.7740 | 0.1557 | |
| PopB-2 | 1091.0731 | 0.0338 | |
| Among SSR alleles within individuals | PopA-1 | 265.0000 | 0.0082 |
| PopA-2 | 501.0000 | 0.0155 | |
| PopA-3 | 231.0000 | 0.0072 | |
| PopB-1 | 239.0000 | 0.0074 | |
| PopB-2 | 146.0000 | 0.0045 | |
| Among populations | 1621.3861 | 0.0503 | |
| Total | 32250.2537 | 1.0000 | |
LD among SSR loci in green foxtail
The extent of LD was assessed among the SSR loci pairs for all accessions, as well as for the subpopulations separately (Table 6). Across all accessions, as many as 60.24% of the total evaluated SSR pairs were in LD (P < 0.05) after Bonferroni correction. For these loci pairs that had significant LD, the value of D′ ranged from 0.237 to 0.93, with a mean of 0.581, and the value of r 2 ranged from 0.004 to 0.092, with a mean of 0.005. The frequency of the pairs of loci with significant LD was reduced by more than half when the LD was calculated within each subpopulation, except for PopB-2 (84.09%). The lowest percentage (11.53) of locus pairs in LD was found in PopB-1. The values of D′ and r 2 were increased when analysed within each subpopulation. PopB-2 presented the highest mean value of 0.950 for D′ and 0.381 for r 2, and the lowest mean value of D′ and r 2 of 0.673 and 0.018 were found in PopA-2.
Table 4.
Molecular diversity of model-based subpopulations inferred by STRUCTURE
| Subpopulations | Sample size | Genotype no./locus | Allele no./locus | Gene diversity/locus | PIC/locus | No. of population- specific alleles |
|---|---|---|---|---|---|---|
| Pop A-1 | 63 | 21.91 | 20.72 | 0.89 | 0.89 | 180 |
| Pop A-2 | 107 | 28.19 | 24.16 | 0.88 | 0.87 | 219 |
| Pop A-3 | 51 | 19.45 | 18.00 | 0.88 | 0.87 | 104 |
| Pop B-1 | 51 | 17.48 | 16.45 | 0.83 | 0.82 | 97 |
| Pop B-2 | 16 | 5.00 | 4.91 | 0.64 | 0.59 | 27 |
Table 6.
Percentage of SSR locus pairs in significant (P < 0.05) LD and LD statistics D′ and r2 of Chinese green foxtail populations
| No. of significant marker pairs in LD | No. of marker pairs evaluated | Fraction of locus pairs (%) | Extent of LD | ||
|---|---|---|---|---|---|
| D′ | r 2 | ||||
| PopA-1 | 128 | 520 | 24.62 | 0.788 | 0.025 |
| PopA-2 | 73 | 597 | 12.23 | 0.673 | 0.018 |
| PopA-3 | 72 | 578 | 12.46 | 0.809 | 0.035 |
| PopB-1 | 101 | 876 | 11.53 | 0.768 | 0.041 |
| PopB-2 | 1274 | 1515 | 84.09 | 0.950 | 0.381 |
| All | 352 | 585 | 60.24 | 0.581 | 0.005 |
Discussion
Molecular diversity of Chinese green foxtail and comparison with that of foxtail millet landraces
This report is the first time that the genetic diversity of Chinese wild green foxtail has been characterized by SSR markers. A majority of the markers (69 of 77) reliably amplified SSRs from the green foxtail accessions, illustrating that green foxtail and foxtail millet are very closely related. This is consistent with the theory that green foxtail is the closet wild ancestor of foxtail millet (Li et al., 1945). Green foxtail and foxtail millet were classified into the same primary gene pool of the AA genome of Setaria (Benabdelmouna et al., 2001a , b ), and they have even been considered to be subspecies of the same species in previous treatments (Dekker, 2003).
The average number of alleles per locus was 33.5 in the Chinese green foxtail, higher than that of 21.4 of the Chinese foxtail millet landraces, using the same set of SSR markers as in previous trials (Wang et al., 2012). This means that a large part of the genetic diversity in the wild gene pool was lost during the domestication of foxtail millet (Supplementary Table S2 at JXB online), and agrees well with previous single nucleotide polymorphism (SNP) analyses of certain genomic regions between these two species (Wang et al., 2010). A higher number of private alleles and lower allele frequencies were also observed in green foxtail compared with foxtail millet landraces (Supplementary Figs S2, S3). This observation is consistent with research conducted on other domesticated crop species (Vigouroux et al., 2002; Kuroda et al., 2006), and indicates the necessity for germplasm collection and protection of the wild relative of crops. The high genetic diversity of green foxtail could provide important genetic resources for foxtail millet improvement programmes and for functional genomics study. Foxtail millet and green foxtail are fast being developed as models of functional genomic investigations for plant morphology and physiology (Doust et al., 2009) and C4 photosynthesis (Brutnell et al., 2010) owing to their small genomes, inbreeding nature, and efficient operation of transformation (Bennetzen et al., 2012). The high genetic diversity of green foxtail is favourable for genetic marker development, construction of segregating populations, functional gene cloning, and association mapping.
Genetic structure of Chinese green foxtail and its geographical distribution
In previous studies of foxtail millet landraces, four subpopulations in China were described, and the genetic structure of the subpopulations was in concordance with the geographical distribution of eco-regions (Wang et al., 2012). Two clusters of green foxtail were clearly identified in this trial, but the distribution of the samples from each cluster was inconsistent with the geographical eco-regions. Nevertheless, PopA includes accessions mainly from higher latitude eco-regions in Northern China, and PopB contained a majority of lines from lower latitude eco-regions in Southern China. A lack of geographical population structure for 22 Asian and European green foxtail accessions was also reported by Le Thierry d’Ennequin et al. (2000) with AFLP markers, although the samples were collected from a much wider geographical range. Some regional geographical structures were identified by ISSR markers (Li et al., 2012), but the geographical distribution structure of 34 green foxtail accessions of world-wide origin was not clear (Li et al., 2012). The sample size of other reports on the green foxtail population structure was too small to warrant discussion (Wang et al., 2010; Hirano et al., 2011).
Why do foxtail millet landraces exist in a clear geographical population structure, while green foxtail accessions do not? Samples of both foxtail millet landraces and green foxtail were collected from the same eco-regions in China, and both kinds of samples were under the same natural environmental conditions of temperature, light, rainfall, and other factors. However, one of the main differences between wild green foxtail and domesticated foxtail millet is the human artificial selection on the cultivated forms, making it probable that human selection was the main factor which created the population structure of Chinese foxtail millet landraces, although this conjecture needs more work in order to be verified.
Wild samples from the vicinity in the same eco-regions probably share the same ancestor and are under the same natural environmental selection, so the geographical population structure in the wild species is a natural phenomenon (Darwin, 1859). The mixed geographical distribution of green foxtail genetic clusters could be due to a variety of factors, including germplasm migration induced by human and animal activities and natural factors; however, the present data are not sufficient to provide a precise cause for this phenomenon.
Foxtail millet origin analyses using green foxtail as reference
As one of the oldest cereals in Eurasia, the origin and domestication of foxtail millet have been of great interest. Studies on morphological diversity (Li et al., 1995; Ochiai et al., 1996), isozyme type (Croullebois et al., 1989), and DNA markers of different kinds (Le Thierry d’Ennequin et al., 2000; Fukunaga et al., 2002, 2006; Li et al., 2012) have all indicated that the highest level of genetic diversity was found in Chinese samples, both of green foxtail and of foxtail millet. The earliest archaeological evidence to date is also located in Northern China (Barton et al., 2009; Lu et al., 2009; Yang et al., 2012). The genetic structure and diversity of Chinese foxtail millet landraces have been investigated, and it has been confirmed that foxtail millet probably originated in the Yellow River regions, where the highest genetic diversity of this species was preserved (Wang et al., 2012). In this trial, high gene diversity and PIC values of green foxtail from Northern China were found, and AMOVAs suggested that Northern China preserved a much higher diversity of green foxtail than other regions. Combining all those data and the earliest archaeological evidence found in the Yellow River region, it can be stated that Northern China is the first domestication centre of foxtail millet, if not the only one.
Understanding the population and geographical structure of both the wild and domesticated types is key in studying domestication. A typical example of this was the identification of Guangxi, China, as the place where rice was domesticated (Huang et al., 2012). The Neighbor–Joining phylogenetic tree constructed using the foxtail millet landraces and green foxtail SSR data (Supplementary Fig. S5 at JXB online) clearly divided the samples into the wild and domesticated gene pools, and four accessions of green foxtail from North China were closely related to foxtail millet, suggesting the origin of Chinese foxtail millet from the northern region. However, the exact place where it was domesticated is still ambiguous, because the four green foxtails that are closely related to foxtail millet are geographically separated. The mixed distribution of genetic clusters in the geographical eco-regions found in this study makes it difficult to answer this question. This is a similar conclusion to those reached by other studies (Le Thierry d’Ennequin et al., 2000; Li et al., 2012). Further genome sequence data and the identification of domestication-related genes would accelerate the understanding of this complex network of lineages of foxtail millet.
Population diversification and gene flow between green foxtail and foxtail millet
Although previous studies on intraspecific hybridization between green foxtail and foxtail millet have indicated repeated genetic introgression (Darmency et al., 1987; Jarvis and Hodgkin, 1999; Wang et al., 1995;Wang et al., 2010), the Neighbor–Joining phylogenetic tree of Chinese green foxtail and foxtail millet showed a clear division between the two gene pools (Supplementary Fig. S5 at JXB online), which suggests that the genetic introgression between the two gene pools is not so frequent. To investigate gene migrations between green foxtail and foxtail millet, Nm was calculated (Slatkin et al., 1989) by conducting classical F ST analyses using previously published data (Wang et al., 2012) (Supplementary Fig. S4). The highest level of gene flow was identified within the green foxtail subpopulations, and the lowest level was characterized between the green foxtail and the foxtail millet landraces (Supplementary Fig. S4). This is consistent with the relatively high level of allele heterozygosity of 0.076 (0.0111–0.6469) identified in this trial for green foxtail (Table 2), while that of foxtail millet was close to zero (Wang et al., 2012). The relatively high level of genetic introgression within the green foxtail millet subclusters may be one of the reasons for the mixed geographical population structure found in this report. The homozygosity of wild green foxtail is lower than that of domesticated foxtail millet, but it is more homozygous than wild rice (Gao et al., 2002; Zhou et al., 2003).
An F-test between green foxtail and foxtail millet revealed 24 SSR loci that had significantly (>97.5%) diversified between these two gene pools, owing to the long period of environmental adaptation or morphological selection. Five loci were localized in the gene-coding regions (Supplementary Table S3 at JXB online), which are potentially important genes of diverged metabolic pathways or have played vital roles in foxtail millet domestication.
Low level of LD of green foxtail
Lower LD was detected in this trial of wild green foxtail compared with a previous study on foxtail millet using the same approach (Wang et al., 2012). This was consistent with research comparing genomic regions of green foxtail and foxtail millet (Wang et al., 2010), and was also similar to analyses of wild and cultivated soybeans (Lam et al., 2010). Based on the present data, the LD level within each subpopulation was higher than the LD value of the total accessions, suggesting that population structure does exist in wild green foxtail. The lower level of LD in green foxtail than in foxtail millet may result from a higher rate of cross-pollination found in the homozygosity analysis of the sampled accessions. Rapid decay of LD also provides more opportunities for identification of potential markers/genes in trait association mapping, which can control important agronomical traits in green foxtail.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Determinations of value of K for substructuring. (A and B) Optimal K identified by LnP(D) and delta K of PopA. (C and D) Optimal K identified by LnP(D) and delta K of PopB.
Figure S2. Shared and specific alleles of wild green foxtail and domesticated foxtail millet detected using SSRs.
Figure S3. Allele frequencies of SSR loci in wild green foxtail and cultivated foxtail millet landraces.
Figure S4. Gene flow estimated by Nm inferred from classical F-test within and between green foxtail and foxtail millet.
Figure S5. Unrooted Neighbor–Joining tree of Chinese S. viridis (blue) and S. italica (green). Four accessions of S. viridis (red) from north China (Chaoyang, Wuan, Changli, and Dingxi) were genetically closer to domesticated S. italica.
Table S1. Genetic diversities identified using SSRs in 288 green foxtail accessions.
Table S2. Comparations of genetic diversity between wild green foxtail and cultivated foxtail millet.
Table S3. List of annotated genes co-localized with SSR loci detected as genomic regions under selection.
Acknowledgements
We thank Dr Andrew Doust from the Department of Botany, Oklahoma State University, USA, for discussion and modification of the English of the manuscript. This work was supported by the ‘863’ program of China (2013AA102603), National Sciences Foundation of China (31171560, 30630045), China Agricultural Research System (CARS07-12.5-A02), National Major Special Fund for New Cultivar Breeding of Genetically Modified Organisms of China (2009ZX08009-093B), and Key Technologies R&D Program of China during the 12th Five-Year Plan period (2011BAD06B01-2). We thank those members of National Millet Crops Research and Development System, CARS, for their help with sample collection.
References
- Austin DF. 2006. Foxtail millets (Setaria: Poaceae)—abandoned food in two hemispheres. Economic Botany 60, 143–158. [Google Scholar]
- Barton L, Newsome SD, Chen F, Wang H, Guilderson TP, Bettinger RL. 2009. Agricultural origins and the isotopic identity of domestication in northern china. Proceedings of the National Academy of Sciences, USA 106, 5523–5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benabdelmouna A, Abirached-Darmency M, Darmency H. 2001a Phylogenetic and genomic relationships in Setaria italica and its close relatives based on the molecular diversity and chromosomal organization of 5S and 18S–5.8S–25S rDNA genes. Theoreitcal and Applied Genetics 103, 668–677. [Google Scholar]
- Benabdelmouna A, Shi Y, Abirached-Darmency M, Darmency H. 2001b Genomic in situ hybridization (GISH) discriminates between the A and B genomes in diploid and tetraploid Setaria species. Genome 44, 685–690. [PubMed] [Google Scholar]
- Bennetzen J, Schmutz J, Wang H, et al. 2012. Reference genome sequence of the model plant Setaria. Nature Biotechnology 30, 555–561. [DOI] [PubMed] [Google Scholar]
- Brutnell TP, Wang L, Swartwood K, Goldschmidt A, Jackson D, Zhu X, Kellogg E, Eck JV. 2010. Setaria viridis: a model for C4 photosynthesis. The Plant Cell 22, 2537–2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caemmerer SV, Quick WP, Furbank RT. 2012. The development of C4 rice: current progress and future challenges. Science 29, 1671–1672. [DOI] [PubMed] [Google Scholar]
- Croullebois ML, Barreneche MT, de Cherisey H, Pernes J. 1989. Intraspecific differentiation of Setaria italica (L.) P.B.: study of abnormalities (weakness, segregation distortion, and partial sterility) observed in F1 and F2 generations. Genome 32, 203–207. [Google Scholar]
- Darwin C. 1859. On the origin of species by means of natural selection or the preservation of favored races in the struggle for life. London: J. Murray. [PMC free article] [PubMed] [Google Scholar]
- Dekker J. 2003. Evolutionary biology of the foxtail (Setaria) species-group. In: Inderjit K, ed. Principles and practices in weed management: weed biology and management. Dordrecht: Kluwer Academic Publishers; 65–114. [Google Scholar]
- Doust AN, Kellogg EA, Devos KM, Bennetzen JL. 2009. Foxtail millet: a sequence-driven grass model system. Plant Physiology 149, 137–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ. 1991. DNA protocol for plants CTAB total DNA isolation. In: Hewitt GM, ed. Molecular techniques in taxonomy. Berlin: Springer, 283–293. [Google Scholar]
- Evanno G, Regnaut S, Goudet J. 2005. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology 14, 2611–2620. [DOI] [PubMed] [Google Scholar]
- Fukunaga K, Ichitani K, Kawase M. 2006. Phylogenetic analysis of the rDNA intergenic spacer subrepeates and its implications for the domestication history of foxtail millet, Setaria italica. Theoretical and Applied Genetics 113, 261–269. [DOI] [PubMed] [Google Scholar]
- Fukunaga K, Wang Z, Kato K, Kawase M. 2002. Geographical variation of nuclear genome RFLPs and genetic differentiation in foxtail millet, Setaria italica (L.) P. Beauv. Genetic Resources and Crop Evolution 49, 95–101. [Google Scholar]
- Gao L, Schaal BA, Zhang C, Jia J, Dong Y. 2002. Assessment of population genetic structure in common wild rice Oryza rufipogon Griff. using microsatellite and allozyme markers. Theoretical and Applied Genetics 106, 173–180. [DOI] [PubMed] [Google Scholar]
- Hedrick PW. 1987. Gametic disequilibrium measures: proceed with caution. Genetics 117, 331–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heerwaarden JV Doebley J Briggs WH Glaubitz JC Goodman MM Jesus JD Gonzalez SG Ross-Ibarra J 2011. Genetic signals of origin, spread, and introgression in a large sample of maize landraces. Proceedings of the National Academy of Sciences, USA 108, 1088–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano R, Naito K, Fukunaga K, Watanabe KN, Ohsawa R, Kawase M. 2011. Genetic structure of landraces in foxtail millet (Setaria italica (L.) P. Beauv.) revealed with transposon display and interpretation to crop evolution of foxtail millet. Genome 54, 498–506. [DOI] [PubMed] [Google Scholar]
- Huang X, Kurata N, Wei X, et al. 2012. A map of rice genome variation reveals the origin of cultivated rice. Nature 490, 497–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis DI, Hodgkin T. 1999. Wild relatives and crop cultivars: detecting natural introgression and farmer selection of new genetic combinations in agroecosystems. Molecular Ecology 8, 159–173. 9919705 [Google Scholar]
- Jin L, Lu Y, Xiao P, Sun M, Corke H, Bao J. 2010. Genetic diversity and population structure of a diverse set of rice germplasm for association mapping. Theoretical and Applied Genetics 121, 475–487. [DOI] [PubMed] [Google Scholar]
- Jusuf M, Pernes J. 1985. Genetic variability of foxtail millet (Setaria italica P. Beauv.). Theoretical and Applied Genetics 63, 117–119. [DOI] [PubMed] [Google Scholar]
- Kruger SA, Able JA, Chalmers KJ, Langridge P. 2004. Linkage disequilibrium analysis of hexaploid wheat. In: Plant & animal genomes XII Conference, 10–14 January, Town & Country Convention Center, San Diego, CA: 321. [Google Scholar]
- Kuroda Y, Kaga A, Tomooka N, Vaughan DA. 2006. Population genetic structure of Japanese wild soybean (Glycine soja) based on microsatellite variation. Molecular Ecology 15, 959–974. [DOI] [PubMed] [Google Scholar]
- Lam H, Xu X, Liu X, et al. 2010. Resequencing of 31 wild and cultivated soybean genomes identifies patterns of genetic diversity and selection. Nature Genetics 42, 1053–1059. [DOI] [PubMed] [Google Scholar]
- Lata C, Gupta S, Prasad M. 2012. Foxtail millet: a model crop for genetic and genomic studies in bioenergy grasses. Critical Reviews in Biotechnology 2012 (in press) [DOI] [PubMed] [Google Scholar]
- Le Thierry d’Ennequin M, Panaud O, Toupance B, Sarr A. 2000. Assessment of genetic relationship between Setaria italica and its wild relative S.viridis using AFLP markers. Theoretical and Applied Genetics 100, 1061–1066. [Google Scholar]
- Li HW, Li CH, Pao WK. 1945. Cytogenetical and genetical studies of the interspecific cross between the cultivated foxtail millet, Setaria italica (L.) Beauv. and the green foxtail millet S. viridis L. Journal of the American Society of Agronomy 37, 32–54. [Google Scholar]
- Li P, Brutnell TP. 2011. Setaria viridis and Setaria italica, model genetic systems for the Panicoid grasses. Journal of Experimental Botany 62, 3031–3037. [DOI] [PubMed] [Google Scholar]
- Li W, Zhi H, Wang Y, Li H, Diao X. 2012. Assessment of genetic relationship of foxtail millet with its wild ancestor and close relatives by ISSR markers. Journal of Integrative Agriculture 11, 556–566. [Google Scholar]
- Li Y, Wu S, Cao Y. 1995. Cluster analysis of an international collection of foxtail millet (Setaria italica (L.) P.Beauv.). Euphytica 83, 79–85. [Google Scholar]
- Liu K, Muse SV. 2005. PowerMarker: integrated analysis environment for genetic marker data. Bioinformatics 21, 2128–2129. [DOI] [PubMed] [Google Scholar]
- Lu H, Zhang J, Liu KB, et al. 2009. Earliest domestication of common millet (Panicum miliaceum) in east Asia extended to 10,000 years ago. Proceedings of the National Academy of Sciences, USA 106, 7367–7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maccaferri M, Sanguineti MC, Noli E, Tuberosa R. 2005. Population structure and long-range linkage disequilibrium in a durum wheat elite collection. Molecular Breeding 15, 271–289. [Google Scholar]
- Mather DE, Hyes PM, Chalmers KJ, Eglinton J, Matus I, Richardson K, Vonzitzewitz J, Marquez-Cedillo L, Hearnden P, Pal N. 2004. Use of SSR marker data to study linkage disequilibrium and population structure in Hordeum vulgare. Perspects for association mapping in barley. In: International barley genetics symposium, Brno, Czech Republic: 20–26 June 2004, 302–307. [Google Scholar]
- Mauro-Herrera M, Wang X, Barbier H, Brutnell TP, Devos KM, Doust AN. 2013. Genetic control and comparative genomic analysis of flowering time in Setaria (Poaceae). G3 (Bethesda) 3, 283–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Takezaki N. 1983. Estimation of genetic distances and phylogenetic trees from DNA analysis. Proceedings of the 5th World Congress on Genetics Applied to Livestock Production 21, 405–412. [Google Scholar]
- Ochiai Y. 1996. Variation in tillering and geographical distribution of foxtail millet (Setaria italica P.Beauv.). Breeding Science 46, 143–146. [Google Scholar]
- Oosterhout CV, Hutchinson WF, Wills DPM, Shipley P. 2004. Micro-checker: software for identifying and correcting genotyping errors in microsatellite data. Molecular Ecology Notes 4, 535–538. [Google Scholar]
- Peakall R, Smouse PE. 2006. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes 6, 288–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stevens M, Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics 155, 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatkin M, Barton NM. 1989. A comparison of three indirect methods for estimating average levels of gene flow. Evolution 43, 1349–1368. [DOI] [PubMed] [Google Scholar]
- Spooner DM, Mclean K, Ramsay G, Waugh R, Bryan GJ. 2005. A single domestication for potato based on multilocus amplified fragment length polymorphism genotyping. Proceedings of the National Academy of Sciences, USA 102, 14694–14699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Molecular Biology and Evolution 24, 1596–1599. [DOI] [PubMed] [Google Scholar]
- Vavilov NL. 1926. Studies on the origin of cultivated plants. Bulletin of Applied Botany and Plant Breeding 26, 1–248. [Google Scholar]
- Vigouroux Y, McMullen M, Hittinger CT, Houchins K, Schulz L, Kresovich S, Matsuoka Y, Doebley J. 2002. Identifying genes of agronomic importance in maize by screening microsatellites for evidence of selection during domestication. Proceedings of the National Academy of Sciences, USA 99, 9650–9655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Chen J, Zhi H, Yang L, Li W, Wang Y, Li H, Zhao B, Chen M, Diao X. 2010. Population genetics of foxtail millet and its wild ancestor. BMC Genetics 11, 90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Jia G, Zhi H, et al. 2012. Genetic diversity and population structure of Chinese foxtail millet [Setaria italica (L.) Beauv.] landraces. G3 (Bethesda) 2, 769–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RL, Wendel JF, Dekker JH. 1995. Weedy adaptation in Setaria spp. 1. Isozyme analysis of genetic diversity and population genetic structure in Setaria viridis. American Journal of Botany 82, 308–317. [Google Scholar]
- Weir BS. 1996. Genetic data analysis II. Sunderland, MA: Sinaur. [Google Scholar]
- Yang X, Wan Z, Perry L, et al. 2012. Early millet use in northern china. Proceedings of the National Academy of Sciences, USA 109, 3726–3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Liu X, Quan Z, et al. 2012. Genome sequence of foxtail millet (Setaria italica) provides insights into grass evolution and biofuel potential. Nature Biotechnology 30, 549–554. [DOI] [PubMed] [Google Scholar]
- Zhou H, Xie Z, Ge S. 2003. Microsatellite analysis of genetic diversity and population genetic structure of a wild rice (Oryza rufipogon Griff.) in China. Theoretical and Applied Genetics 107, 332–339. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.