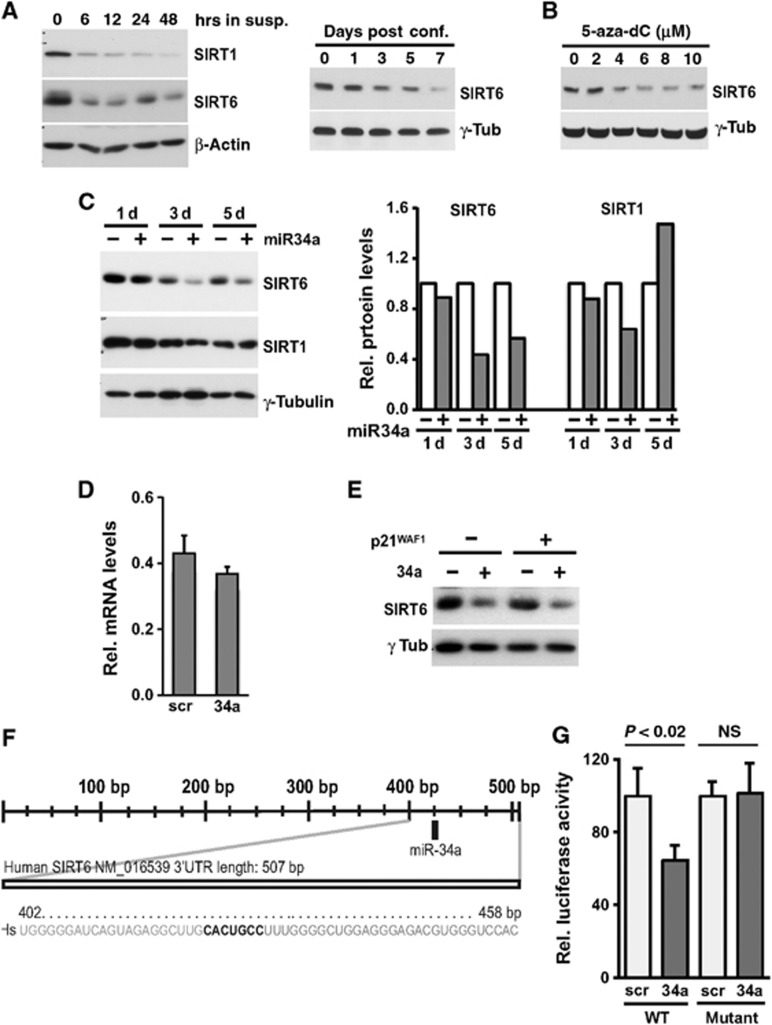
Figure 7.
SIRT6 is a miR-34a target downregulated during differentiation. (A) HKCs were induced to differentiate either by culture in suspension or high confluency conditions for the indicated times (left and right panels, respectively) and levels of SIRT1 and SIRT6 expression were determined by immunoblot analysis with β-actin and γ-tubulin, respectively, as equal loading controls. (B) SCC13 cells were treated with the indicated concentrations (μM) of 5-aza-2′-deoxycytidine for 4 days and levels of SIRT6 expression were determined by immunoblot analysis with γ-tubulin as equal loading control. (C) HKCs were transfected with miR-34a precursor (+) or scrambled control oligonucleotides (−) for the indicated times (days) followed by immunoblot analysis of the indicated proteins. SIRT1 and SIRT6 blots were performed on parallel gel/blot by sequential blotting of the same membrane without stripping with γ-tubulin giving the same pattern of expression (left panel). Relative protein levels were quantified after densitometric scanning of the autoradiographs and normalization for γ-tubulin expression (right panel). (D) HKCs were transfected for 3 days with miR-34a precursor (+) or scrambled control oligonucleotides (−) and analysed for levels of SIRT6 mRNA by qRT–PCR. (E) HKCs stably infected with a lentivirus for doxycycline-inducible expression of p21WAF1/Cip1 were either untreated (−) or treated with doxycycline (500 ng/ml) for 5 days (+). Cells were transfected with miR-34a mimics (25 nM) or scrambled controls for the last 3 days of the experiment followed by immunoblot analysis of SIRT6 expression with γ-tubulin as equal loading control. (F) The human SIRT6 3′-UTR was analysed for the presence of miR-34a recognition sites by Targetscan 5.2 and Miranda softwares. Shown in bold is the position of the miR-34a binding site in the human SIRT6 3′-UTR. Numbers refer to the nucleotide position starting from the stop codon of the SIRT6 coding region. (G) HKCs were co-transfected in triplicate wells with a luciferase reporter construct with the WT SIRT6-3′UTR sequence or one lacking the miR-34a binding site (mutant) together with miR-34a precursor or scrambled control oligonucleotides. Luciferase activity was measured 48 h later, with total protein levels for normalization.
Source data for this figure is available on the online supplementary information page.