Abstract
Pre-eclampsia is a common vascular disorder of pregnancy. It originates in the placenta and targets the maternal endothelium. According to epidemiological research, >50% of the liability to this disorder can be accounted for by genetic factors. Both maternal and fetal genes contribute to the risk, but especially the fetal genetic risk profile is still poorly understood. We have previously detected linkage signals in multiplex Finnish families on chromosomes 2p25, 4q32, and 9p13 using maternal phenotypes. We performed a linkage analysis using updated maternal phenotypes and an unprecedented linkage analysis using fetal phenotypes. Markers genotyped were available from 237 individuals in 15 Finnish families, including 72 affected mothers and 49 affected fetuses. The MERLIN software was used for sample and marker quality control and linkage analysis. The results were compared against the original ones obtained by using the GENEHUNTER 2.1 software. The previous identification of the maternal susceptibility locus to a genetic location at 21.70 cM near marker D2S168 on chromosome 2 was confirmed by using both maternal and fetal phenotypes (maternal non-parametric linkage (NPL) score 3.79, P=0.00008, LOD (logarithm (base 10) of odds)=2.20 and fetal NPL score 2.95, P=0.002, LOD=1.71). As a novel finding, we present a suggestive linkage to chromosome 18 at 86.80 cM near marker D18S64 (NPL score 2.51, P=0.006, LOD=1.20) using the fetal phenotype. We propose that chromosome 18 may harbor a new fetal susceptibility locus for pre-eclampsia.
Keywords: pre-eclampsia, linkage, maternal phenotype, fetal phenotype, family study
Introduction
Pre-eclampsia is a common disorder of pregnancy that increases maternal and fetal morbidity.1 In the Western world, the organized prenatal care has decreased maternal mortality, but the risk is often passed on to the fetus in the form of a premature birth.2 Pre-eclampsia has characteristics similar to vascular disorder, such as reduced placental perfusion and hypertension, along with endothelial dysfunction and proteinuria.1 Clinical observations suggest that early-onset and late-onset disease types can be separated in origin, even though individual cases share characteristics.3 The progression of the disease is unpredictable and may lead to a convulsive phase, eclampsia. The removal of the affected placenta through delivery is the only known treatment for pre-eclampsia.
The disease is associated with a risk of later-life conditions, such as cardiovascular diseases and diabetes, in both the mother who has gone through a pre-eclamptic pregnancy and the child born of a pre-eclamptic pregnancy.4 Although pre-eclampsia is a highly complex disease with both environmental and genetic factors, the inherited element accounts for >50% of an individual's disease susceptibility.5 Epidemiological research has demonstrated that both maternal and fetal genes are important in the pathogenesis.6
Genome-wide linkage and association studies carried out by us7 and others8, 9, 10, 11, 12, 13 have revealed a number of maternal susceptibility loci. The placental origin of pre-eclampsia suggests a central role for the fetal genotype in the pathogenesis, and the role of the maternal–fetal gene interaction is supported by twin studies.14 The results yielded by candidate gene studies have, thus far, proved inconsistent and addressed mainly the maternal genotype and the susceptibility loci therein.4, 9, 10, 11, 12, 13
In this follow-up study, we make comparisons of the earlier and more recent methods regarding the linkage findings in pre-eclampsia. In particular, we wish to shed light on the assumed genetic connection between the maternal and fetal pre-eclamptic phenotypes.
Materials and methods
In our original linkage analysis on 15 multiplex pre-eclampsia families, the patients with pre-eclampsia were recruited predominantly from the Kainuu province in central eastern Finland and in the Helsinki region in southern Finland. Their families were ascertained on the basis of at least one proteinuric pre-eclampsia case. Within these families, women who had gestational hypertension without proteinuria were also considered as affected because of the higher likelihood of shared susceptibility genes within families.
In the original study, we found two loci exceeding the threshold for significant linkage: chromosome 2p25 and 9p13.7 In addition, there was a locus showing suggestive linkage at chromosome 4q32. In this follow-up study, we performed a non-parametric linkage (NPL) analysis using the same sample set and microsatellite markers as reported in the original linkage study.
We defined an affected mother as having either pre-eclampsia or gestational hypertension in the pregnancy, and an affected fetus as an individual born of a pregnancy affected by either of these. The clinical criteria for pre-eclampsia were defined as a systolic blood pressure >140 mm Hg or a diastolic blood pressure >90 mm Hg in a woman who was normotensive before 20 weeks of gestation and an urinary protein excretion of ≥0.3 g in 24-h sample. Without the proteinuria, the same criteria account for a gestational hypertension (GHT) diagnosis.
A total of 237 persons from 15 Finnish families gave samples, of which72 were considered as affected mothers (cases of pre-eclampsia n=54, eclampsia n=1 and GHT n=17), and 49 affected fetuses (born of a pre-eclamptic pregnancy n=45 and of a GHT pregnancy n=4). The MERLIN software15 was used for sample and marker quality control and linkage analysis. Results were compared against the original ones obtained by using the GENEHUNTER 2.1 software. The complex nature of pre-eclampsia discourages the assumption of a fixed pattern of heritability. Therefore, and to produce optimally commensurate analysis, a non-parametric approach was chosen.
Results
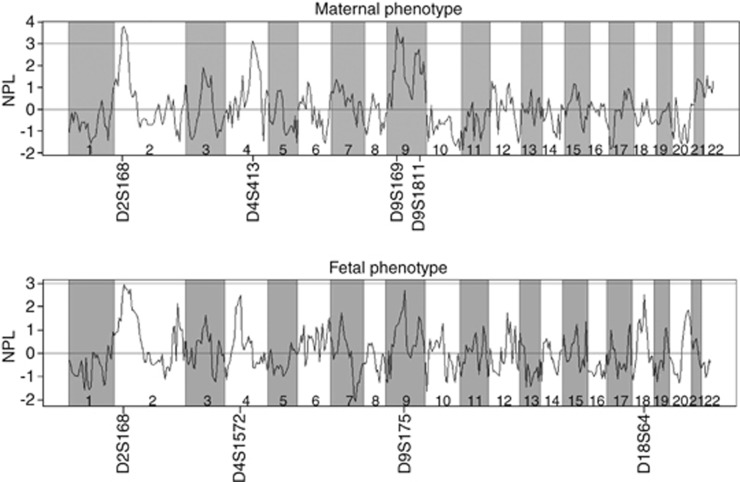
The peaks supporting significant (NPL score>3, P<0.0001) or suggestive (NPL score>2, P=0.0001–0.01) linkage as defined by the standard set by Kruglyak and Lander,16 remained roughly in the same locations as on the chromosomes of the original genome scan. Genome-wide multipoint NPL analysis results for both maternal and fetal phenotypes are presented in Figure 1.
Figure 1.
Multipoint NPL analysis results of genome wide-scale for both maternal and fetal phenotypes.
Fetal and maternal NPL score diagrams correlated well on chromosome 2. The maximum NPL scores on chromosome 2 were at 21.70 cM near marker D2S168 (NPL score 3.79, P=0.00008, LOD=2.20) using the maternal phenotype and (NPL score 2.95, P=0.002, LOD=1.71) using the fetal phenotype. On chromosome 4, the highest peak was at 158.20 cM near marker D4S413 (NPL score 3.13, P=0.0009, LOD=2.09) using the maternal phenotype and at 109.60 cM near marker D4S1572 (NPL score 2.50, P=0.006, LOD=0.78) using fetal phenotype. On chromosome 9 the highest peak using the maternal phenotype was at 38.90 cM near marker D9S169 (NPL=3.76, P=0.00008, LOD=1.31), and on the other side of the chromosome, at 120.80 cM near marker D9S1811 (NPL=2.74, P=0.003, LOD=1.29). The maximum peak on chromosome 9 using the fetal phenotype was found at 59.20 cM near marker D9S175 (NPL=2.69, P=0.004, LOD=1.22). Although lacking a counterpart in the maternal phenotype, the highest peak on the fetal chromosome 18 suggested linkage at 86.80 cM, near marker D18S64 (NPL score of 2.51, P=0.006, LOD=1.20).
In addition to these nominal P-values, we also used Merlin to run simulations. The simulated cases with P-values and NPL scores superior to ones obtained in analyzing maternal chromosomes 2, 4 and 9 stayed under the threshold of 0.01. In chromosomes 2 and 4 of the fetal phenotype, threshold of 0.05 was attained, although in chromosomes 9 and 18, this threshold was exceeded. The locations of linkage in this study compared with the original study are collectively presented in Table 1.
Table 1. Locations of suggested and significant linkage in Finnish pre-eclampsia families, according to phenotypes.
| NPL score in the original studya |
NPL score in the follow-up study using Merlin |
||||
|---|---|---|---|---|---|
| Chromosome | Marker | Location in cM | Maternal | Maternal | Fetal |
| 2 | D2S168 | 21.7 | 3.77 | 3.79 | 2.95 |
| 4 | D4S1572 | 109.6 | 2.50 | ||
| 4 | D4S413 | 158.2 | 3.13 | ||
| 4 | D4S3046 | 163 | 3.13 | ||
| 9 | D9S169 | 38.9 | 3.74 | 3.76 | |
| 9 | D9S175 | 59.2 | 2.69 | ||
| 15 | D15S1007 | 24.5 | 2.00 | ||
| 18 | D18S64 | 86.8 | 2.51 | ||
NPL, non-parametric linkage.
Laivuori et al.7
Discussion
In summary, we confirm the previously identified maternal susceptibility locus to chromosome 2 near marker D2S168 using both maternal and fetal phenotypes. Additionally, we present a suggestive fetal susceptibility locus to chromosome 18 near marker D18S64 as a novel finding.
The markers implying the highest susceptibility to linkage in this study have also been found interesting in other studies, concerning the diseases closely associated with pre-eclampsia. Such diseases are notably diabetes,17 cardio-vascular disorders18 and hypertension.19, 20, 21 The promising findings in the maternal and fetal scan of the chromosome 2 may benefit the search of rare genetic variants in Finnish families. The fine mapping of the chromosome 18 peak region would improve our knowledge of the seldom highlighted fetal phenotype. The linkage similarities between the two sets of phenotypes, on chromosome 2 and to some extent on chromosome 9, encourage further studies emphasizing the role of maternal–fetal interaction. Neither linkage nor genome-wide association studies have thus far succeeded in identifying pre-eclampsia-associated genetic variants. Our results underline the importance of collecting more carefully characterized families in order to improve statistical power. They also signify the role of the families with pre-eclampsia as an important resource for studies using next-generation sequencing methods. The essential genetic interactions yet to be discovered may well prove to be defined by the genes revealed in linkage scans today.
Acknowledgments
We are indebted to all of the Finnish families whose participation made this work possible. We thank Leena Järvinen for technical assistance. This study was supported by grants from the Academy of Finland, the Research Funds of the University of Helsinki, the Government Special Subsidiary for Health Sciences (EVO funding) at Helsinki and Uusimaa Hospital District, the Helsingin Sanomat Centennial Foundation, the Jalmari and Rauha Ahokas Foundation, the Maud Kuistila Foundation, the Uusimaa Regional Fund of the Finnish Cultural Foundation, the Päivikki and Sakari Sohlberg Foundation, the Jane and Aatos Erkko Foundation and the Finnish Medical Foundation.
The authors declare no conflict of interest.
References
- Roberts JM, Taylor RN, Musci TJ, Rodgers GM, Hubel CA, McLaughlin MK. Preeclampsia: an endothelial cell disorder. Am J Obstet Gynecol. 1989;161:1200–1204. doi: 10.1016/0002-9378(89)90665-0. [DOI] [PubMed] [Google Scholar]
- Backes CH, Markham K, Moorehead P, et al. Maternal Preeclampsia and Neonatal Outcomes J Pregnancy 20112011doi: 10.1155/2011/214365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oudejans CBM, van Dijk M, Oosterkamp M, Lachmeijer A, Blankenstein MA. Genetics of preeclampsia. Hum Genet. 2006;120:607–612. doi: 10.1007/s00439-006-0259-1. [DOI] [PubMed] [Google Scholar]
- Chappell S, Morgan L. Searching for genetic clues to the causes of pre-eclampsia. Clin Sci. 2006;110:443–458. doi: 10.1042/CS20050323. [DOI] [PubMed] [Google Scholar]
- Skjaerven R, Vatten LJ, Wilcox AJ, Ronning T, Irgens LM, Lie RT. Recurrence of pre-eclampsia across generations: exploring fetal and maternal genetic components in a population based cohort. BMJ. 2005;331:877. doi: 10.1136/bmj.38555.462685.8F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cnattingius S, Reilly M, Pawitan Y, Lichtenstein P.Maternal and fetal genetic factors account for most of familial aggregation of preeclampsia: a population-based Swedish cohort study Am J Hum Genet 2004. Part A130365–371. [DOI] [PubMed] [Google Scholar]
- Laivuori H, Lahermo P, Ollikainen V, et al. Susceptibility loci for preeclampsia on chromosomes 2p25 and 9p13 in Finnish families. Am J Hum Genet. 2003;72:168–177. doi: 10.1086/345311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison GA, Humphrey KE, Jones N, et al. A genomewide linkage study of preeclampsia/eclampsia reveals evidence for a candidate region on 4q. Am J Hum Genet. 1997;60:1158–1167. [PMC free article] [PubMed] [Google Scholar]
- Lachmeijer AM, Arngrímsson R, Bastiaans EJ, et al. A genome-wide scan for preeclampsia in the Netherlands. Eur J Hum Genet. 2001;9:758–764. doi: 10.1038/sj.ejhg.5200706. [DOI] [PubMed] [Google Scholar]
- Arngrímsson R, Siguróaróóttir S, Frigge ML, et al. A genome-wide scan reveals a maternal susceptibility locus for pre-eclampsia on chromosome 2p13. Hum Mol Genet. 1999;9:1799–1805. doi: 10.1093/hmg/8.9.1799. [DOI] [PubMed] [Google Scholar]
- Moses EK, Lade JA, Guo G, Wilton AN, et al. A genome scan in families from Australia and New Zealand confirms the presence of a maternal susceptibility locus for pre-eclampsia on chromosome 2. Am J Hum Genet. 2000;67:1581–1585. doi: 10.1086/316888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MP, Fitzpatrick E, Dyer TD, et al. Identification of two novel quantitative trait loci for pre-eclampsia susceptibility on chromosomes 5q and 13q using a variance components-based linkage approach. Mol Hum Reprod. 2007;13:61–67. doi: 10.1093/molehr/gal095. [DOI] [PubMed] [Google Scholar]
- Johnson MP, Brennecke SP, East CE, Göring HH, et al. Genome-wide association scan identifies a risk locus for preeclampsia on 2q14, near the inhibin, beta B gene. PLoS One. 2012;7:e33666. doi: 10.1371/journal.pone.0033666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ros Salonen H, Lichtenstein P, Lipworth L, Cnattingius S. Genetic effects on the liability of developing pre-eclampsia and gestational hypertension. Am J Med Genet. 2000;91:256–260. [PubMed] [Google Scholar]
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin–rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- Kruglyak L, Daly MJ, Reeve-Daly M, Lander ES. Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet. 1996;58:1347–1363. [PMC free article] [PubMed] [Google Scholar]
- Merriman T, Twells R. Evidence by allelic association-dependent methods for a type 1 diabetes polygene (IDDM6) on chromosome 18q21. Hum Mol Genet. 1997;6:1003–1010. doi: 10.1093/hmg/6.7.1003. [DOI] [PubMed] [Google Scholar]
- Bellis C, Cox HC, Dyer TD, et al. Linkage mapping of CVD risk traits in the isolated Norfolk Island population. Hum Genet. 2008;124:543–552. doi: 10.1007/s00439-008-0580-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angius A, Petretto E, Battista Maestrale G. A new essential hypertension susceptibility locus on chromosome 2p24-p25, detected by genomewide search. Am J Hum Genet. 2002;71:893–905. doi: 10.1086/342929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu DL, Wang HY, Xiong MM, He X. Linkage of hypertension to chromosome 2q14-q23 in Chinese families. J Hypertens. 2001;19:55–61. doi: 10.1097/00004872-200101000-00008. [DOI] [PubMed] [Google Scholar]
- Kamide K, Kokubo Y, Yang J, Tanaka C. Hypertension susceptibility genes on chromosome 2p24-p25 in a general Japanese population. J Hypertens. 2005;23:955–960. doi: 10.1097/01.hjh.0000166835.70935.3c. [DOI] [PubMed] [Google Scholar]