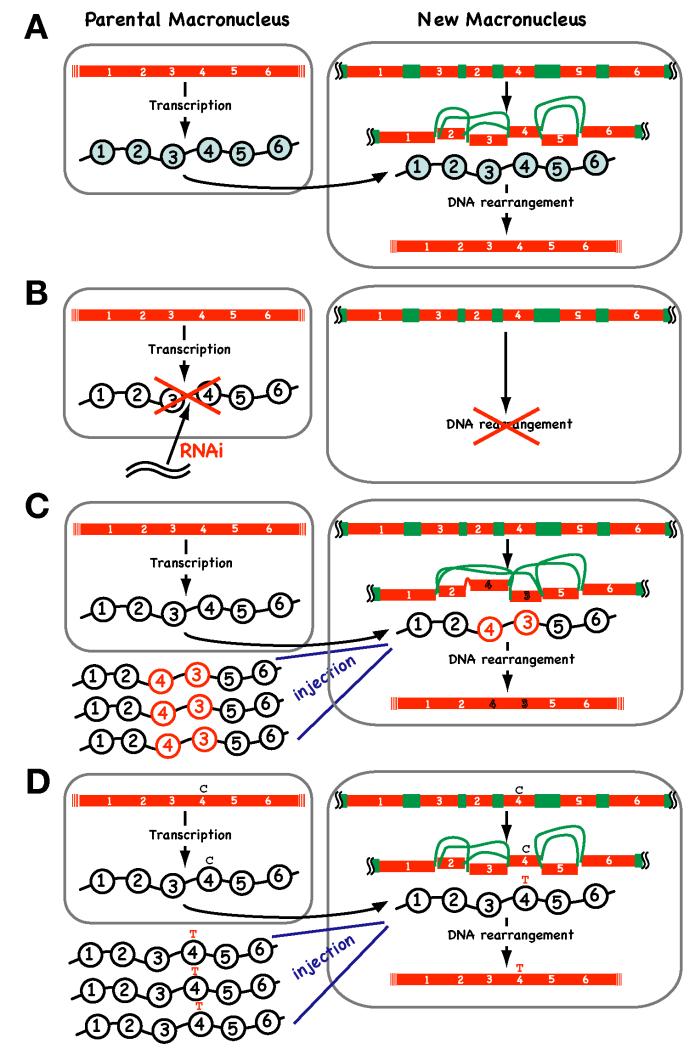
Figure 7. Long non-coding RNA-guided DNA descrambling in Oxytricha.
(A) A model for RNA-guided DNA descrambling in Oxytricha. Telomere-to-telomere transcription of parental macronuclear “gene-sized” chromosomes produces guide RNAs (wavy lines with circles), which are then transported to the newly developed macronucleus where they act as scaffolds to guide DNA rearrangement. (B) Disruption of long non-coding RNAs by RNAi causes a defect in DNA rearrangement. (C) Microinjection of artificial templates (in this example, RNA having a 1-2-4-3-5-6 sequence) alters the order of the DNA descrambling pattern in the new macronucleus. (D) Microinjection of artificial templates that have base substitutions (C to T in this example) alters the DNA sequence of the new macronucleus.