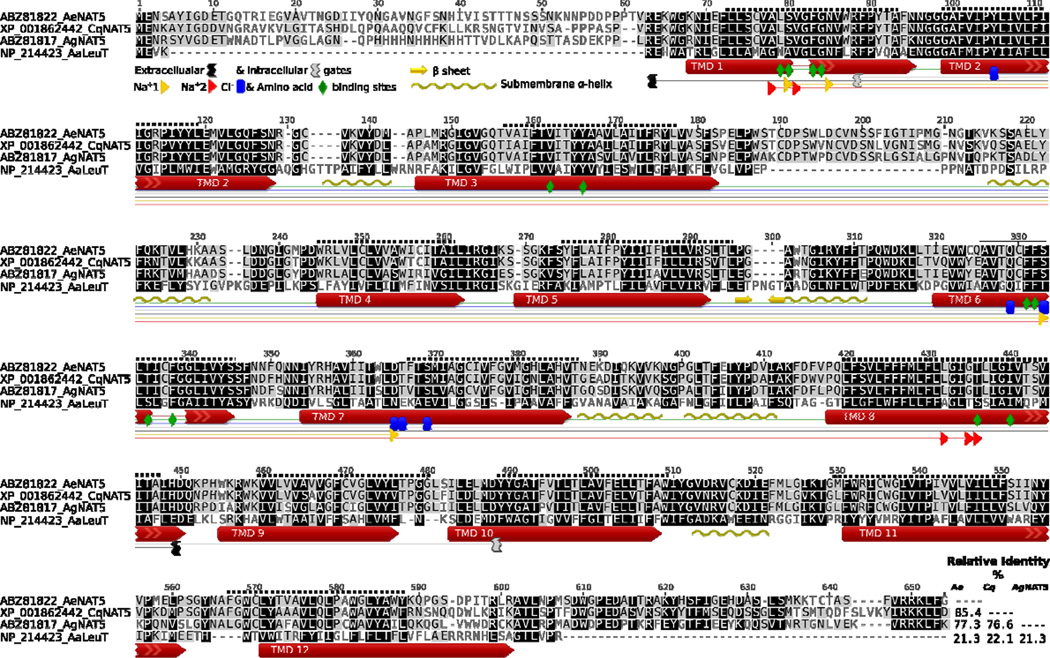
Figure 1. Protein sequence alignment of AeNAT5 and orthologous mosquito NATs versus the sequence and structural motifs of the LeuT.
The background intensity indicates sequence similarity that was rendered using a Blosum 45 matrix and 0 threshold optional settings. Colored shapes show relative positions of specific structural motifs and 12 TMDs in LeuT (long red arrows and specific #, colored shapes legend insert). The uncoiled fragments of TMDs 1 and 6 are shown as lines. The dotted line above the sequence is the relative position of theoretically predicted TMDs in AeNAT5. The graphical annotation of aligned positions and inserts demonstrates 12 amino acid binding sites, 4 and 5 sites coordinating the 1st and 2nd Na+ ions, 5 putative sites coordinating Cl−, and the charged residues of intracellular and extracellular gates of the transporters. The beta sheet hairpin and sub-membrane helices identified in the LeuT structure are indicated by yellow arrows and wave lines, respectively. The right corner insert displays a pairwise identity matrix (%) for aligned NATs without N and C termini.